NMR Spectroscopy for Protein Higher Order Structure Similarity Assessment in Formulated Drug Products
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Review of Current Real-World Experience with Teriparatide As Treatment of Osteoporosis in Different Patient Groups
Journal of Clinical Medicine Review Review of Current Real-World Experience with Teriparatide as Treatment of Osteoporosis in Different Patient Groups Barbara Hauser 1,2,* , Nerea Alonso 2 and Philip L Riches 1,2 1 Rheumatic Disease Unit, Western General Hospital, NHS Lothian, Edinburgh EH4 2XU, UK; [email protected] 2 Rheumatology and Bone Disease Unit, Centre for Genomic and Experimental Medicine, MRC Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh EH4 2XU, UK; [email protected] * Correspondence: [email protected] Abstract: Teriparatide has proven effective in reducing both vertebral and non-vertebral fractures in clinical trials of post-menopausal and glucocorticoid-induced osteoporosis. Widespread adoption of Teriparatide over the last two decades means that there is now substantial experience of its use in routine clinical practice, which is summarized in this paper. Extensive real-world experience of Teriparatide in post-menopausal osteoporosis confirms the fracture and bone density benefits seen in clinical trials, with similar outcomes identified also in male and glucocorticoid-induced osteoporosis. Conversely, very limited experience has been reported in pre-menopausal osteoporosis or in the use of Teriparatide in combination with other therapies. Surveillance studies have identified no safety signals relating to the possible association of Teriparatide with osteosarcoma. We also review the evidence for predicting response to Teriparatide in order to inform the debate on where best to use Teriparatide in an increasingly crowded therapeutic landscape. Citation: Hauser, B.; Alonso, N.; Riches, P.L. Review of Current Keywords: Teriparatide; anabolic treatment; osteoporosis; fracture Real-World Experience with Teriparatide as Treatment of Osteoporosis in Different Patient Groups. -

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

Novel Therapies in Osteoporosis: PTH-Related Peptide Analogs and Inhibitors of Sclerostin
62 2 Journal of Molecular T D Rachner et al. Novel anabolic osteoporosis 62:2 R145–R154 Endocrinology treatments REVIEW Novel therapies in osteoporosis: PTH-related peptide analogs and inhibitors of sclerostin Tilman D Rachner1,2,3, Lorenz C Hofbauer1,2,3,4, Andy Göbel1,3 and Elena Tsourdi1,2,3 1Department of Medicine III, Technische Universität Dresden Medical Center, Dresden, Germany 2Center for Healthy Aging, Technische Universität Dresden Medical Center, Dresden, Germany 3German Cancer Consortium (DKTK), partner site Dresden and German Cancer Research Center (DKFZ), Dresden, Germany 4Center for Regenerative Therapies Dresden, Technische Universität Dresden, Dresden, Germany Correspondence should be addressed to T D Rachner: [email protected] Abstract Bone-forming approaches to treat patients with severe osteoporosis are effective, but Key Words treatment options are limited, and there is an unmet clinical need for additional drugs. f PTH This review discusses two novel and advanced anabolic therapeutic concepts that f PTH-related protein have successfully completed phase 3 trials. Romosozumab is a monoclonal antibody f abaloparatide that targets the Wnt inhibitor sclerostin. Two phase 3 trials (FRAME and ARCH) of f sclerostin romosozumab for the treatment of postmenopausal osteoporosis have been completed. f sclerostin antibody Both trials successfully reached their primary endpoint by reducing vertebral fractures by f romosozumab 75% compared to placebo (FRAME trial) and 48% compared to alendronate (ARCH trial), respectively. Abaloparatide is a PTH-related protein (PTHrP) analog that has displayed bone anabolic activity. In the phase 3 ACTIVE trial, abaloparatide was compared to placebo and teriparatide for 18 months in postmenopausal women who had already experienced an osteoporotic fracture. -

AACE Annual Meeting 2021 Abstracts Editorial Board
June 2021 Volume 27, Number 6S AACE Annual Meeting 2021 Abstracts Editorial board Editor-in-Chief Pauline M. Camacho, MD, FACE Suleiman Mustafa-Kutana, BSC, MB, CHB, MSC Maywood, Illinois, United States Boston, Massachusetts, United States Vin Tangpricha, MD, PhD, FACE Atlanta, Georgia, United States Andrea Coviello, MD, MSE, MMCi Karel Pacak, MD, PhD, DSc Durham, North Carolina, United States Bethesda, Maryland, United States Associate Editors Natalie E. Cusano, MD, MS Amanda Powell, MD Maria Papaleontiou, MD New York, New York, United States Boston, Massachusetts, United States Ann Arbor, Michigan, United States Tobias Else, MD Gregory Randolph, MD Melissa Putman, MD Ann Arbor, Michigan, United States Boston, Massachusetts, United States Boston, Massachusetts, United States Vahab Fatourechi, MD Daniel J. Rubin, MD, MSc Harold Rosen, MD Rochester, Minnesota, United States Philadelphia, Pennsylvania, United States Boston, Massachusetts, United States Ruth Freeman, MD Joshua D. Safer, MD Nicholas Tritos, MD, DS, FACP, FACE New York, New York, United States New York, New York, United States Boston, Massachusetts, United States Rajesh K. Garg, MD Pankaj Shah, MD Boston, Massachusetts, United States Staff Rochester, Minnesota, United States Eliza B. Geer, MD Joseph L. Shaker, MD Paul A. Markowski New York, New York, United States Milwaukee, Wisconsin, United States CEO Roma Gianchandani, MD Lance Sloan, MD, MS Elizabeth Lepkowski Ann Arbor, Michigan, United States Lufkin, Texas, United States Chief Learning Officer Martin M. Grajower, MD, FACP, FACE Takara L. Stanley, MD Lori Clawges The Bronx, New York, United States Boston, Massachusetts, United States Senior Managing Editor Allen S. Ho, MD Devin Steenkamp, MD Corrie Williams Los Angeles, California, United States Boston, Massachusetts, United States Peer Review Manager Michael F. -

2017 Fda Peptide Harvest
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 10 April 2018 doi:10.20944/preprints201804.0126.v1 Peer-reviewed version available at Pharmaceuticals 2018, 11, 42; doi:10.3390/ph11020042 1 Review 2 2017 FDA PEPTIDE HARVEST 3 Othman Al Musaimi,1,2,# Danah Alshaer, 1,2,# Beatriz G. de la Torre,3,* Fernando Albericio,2,4,5.* 4 1 College of Health Sciences, University of KwaZulu-Natal, Durban 4000, South Africa 5 2 School of Chemistry, University of KwaZulu-Natal, Durban 4001, South Africa 6 3 KRISP, College of Health Sciences, University of KwaZulu-Natal, Durban 4001, South Africa 7 4 CIBER-BBN, Networking Centre on Bioengineering, Biomaterials and Nanomedicine, University of 8 Barcelona, 08028 Barcelona, Spain 9 5 Department of Organic Chemistry, University of Barcelona, 08028 Barcelona, Spain 10 * Correspondence: [email protected]; [email protected]; Tel.: +27-614009144 11 12 13 Abstract: 2017 was an excellent year in terms of new drugs (chemical entities and biologics) 14 approved by the FDA, with a total of forty-six. In turn, one of the highlights was the number of 15 peptides (six) included in this list. Here, the six peptides are analysed in terms of chemical structure, 16 synthetic strategy used for their production, source, biological target, and mode of action. 17 Keywords: pharmaceutical market; drugs; drug discovery; solid-phase peptide synthesis 18 Introduction 19 The financial investment associated with the pharmaceutical industry is one of the largest in the 20 industrial sector—surpassed only by the telecommunications sector. However, the number of new 21 products (drugs) entering the market each year is relatively low. -

The European Pharmacopoeia (Ph. Eur.)
THE EUROPEAN DIRECTORATE FOR THE QUALITY OF MEDICINES & HEALTHCARE (EDQM) European Directorate for the Quality of Medicines & HealthCare • A Council of Europe Directorate, based on the Convention on the Elaboration of a European Pharmacopoeia (PA, 1964) • Mission: to contribute to a basic human right: access to good quality medicines and healthcare • See www.edqm.eu for further information 2 ©2020 EDQM, Council of Europe. All rights reserved. Key players for the quality of medicines in Europe National Authorities EU Coordination of scientific DG Health and resources from MS Food Safety Licensing Authorities Inspectorates Control Laboratories Pharmacopoeia Authorities Pharmaceutical legislation National Authorities EU & non-EU European Pharmacopoeia OMCL, Certification, Healthcare…. 3 ©2020 EDQM, Council of Europe. All rights reserved. •EDQM Laboratory •Established in 1967 •Located in Strasbourg, France • - Analytical Chemistry Division • - Biological Section 4 ©2020 EDQM, Council of Europe. All rights reserved. Role of the EDQM Laboratory European • establish/monitor Ph. Eur. reference substances (CRS) Pharmacopoeia • support elaboration of Ph. Eur. monographs / chapters (Ph. Eur.) • support the biological standardisation programme European • support the annual PTS programme OMCL network WHO • establish/monitor ICRS (Ref. Standards for the Ph. Int.) • establish/monitor ISA (Int’l Standards for Antibiotics) • participate in int’l studies on RS 5 ©2020 EDQM, Council of Europe. All rights reserved. European Pharmacopoeia Reference Standards for Biologicals Frank Jung, Ph.D. EDQM Laboratory Department Biological Section 6 ©2020 EDQM, Council of Europe. All rights reserved. The European Pharmacopoeia (Ph. Eur.) A compendium of quality standards for substances for pharmaceutical use and medicinal products Legally binding in 39 countries signatories of the Ph. -

Nitroglycerin Ointment Strengthened Bone
16 OSTEOPOROSIS MARCH 2011 • CLINICAL ENDOCRINOLOGY NEWS Nitroglycerin Ointment Strengthened Bone BY MARY ANN MOON cerin or a matching placebo ointment to included in the analysis: 126 in the ni- The primary end point was change in a piece of onion skin that was taped to the troglycerin group and 117 in the placebo lumbar spine areal BMD after 2 years. FROM JAMA upper outer arm overnight, every night. group. A total of 106 subjects dropped Those who received active nitroglycerin opical nitroglycerin ointment rais- The study subjects were women aged 50 out because of headache, nausea, or al- showed a significant increase of about es bone mineral density, cuts re- years or older (mean age, 62 years) who lergic reaction, and another 51 “lost in- 7%, compared with women in the place- Tsorption, and alters bone struc- were at least 1 year past menopause. terest” or became ineligible. bo group, in that measure. ture so that bone strength is increased, None had osteoporosis, but all had BMD After randomization, another 30 sub- They also showed comparable increas- according to results of a double-blind T scores of 0 to –2.0 at the lumbar spine jects in the treatment group and 15 in the es in areal BMD at the total hip (6%) and trial in 243 women. and higher than –2.0 at the total hip. placebo group discontinued or were lost femoral neck (7%). Compared with place- The magnitude of improvement equals Of 400 women enrolled, only 243 re- to follow-up, including 26 who cited ad- bo users, the nitroglycerin group also or exceeds that observed with other ther- mained in the study long enough to be verse reactions including headache. -

Moon Jung, Waters
A Systematic Approach for Improving the Recovery of Hydrophobic Peptides during LC-MS Analyses November 2019 Moon Chul (Moon) Jung, Ph.D. Waters Corporation, Chemistry Technology Center R&D ©2019 Waters Corporation COMPANY CONFIDENTIAL 1 Proteins and peptides can be quite sticky! Molecule or ¡ Non-Specific Binding (NSB) or Non-Specific Adsorption (NSA) Surface – Biomolecules tend to adhere to any exposed surfaces. – Any chemical interaction can be the source of NSB, Non-polar Polar but most dominantly… Uncharged Charged Polar Polar o Polarity-based interactions, e.g., hydrophobic attraction o Ionic interactions, e.g., coulombic attraction Basic Acidic (cationic) (anionic) ¡ NSB of biomolecules is more difficult to deal with compared to NSB of small molecules – Biomolecules are larger and more complex than small molecules. o There may be multiple binding interactions between biomolecules. – Proteins may be cooperatively deformed during the adsorption process. o And may be permanently lost. ©2019 Waters Corporation COMPANY CONFIDENTIAL Figure from F. Poncin et al., J. Func. Biomater. 3(3), 528, 2012 2 How does Non-Specific Binding (NSB) affect analyses? 5 ng/mL [10µL] - WithoutLeuprolide + RP Dilution - 95:5 (MW- Waters Untreated1209.4) - Rep 1 hormone antagonist peptide 100 50x scale Technical Replicate 1 Minimal NSB 5 ng/mL [10µL] - Without + RP Dilution - 95:5 - Waters Untreated - Rep 1 100 Technical Replicate 2 CV: 1.7% Technical Replicate 3 (n = 3) % CV: 41.8% % (n = 3) Non-specific binding can lead to: – Low sensitivity – High variability -

Scibx Highlights
ANALYSIS FROM THE MAKERS OF AND NOvember 17, 2011 • vOLUme 4 / NUmber 45 THIS WEEK ANALYSIS Dampening COVER STORY 1 Dampening neuroinflammation A California team has shown that inhibiting MAGL reduces neuroinflammation neuroinflammation in a mouse model of Parkinson’s disease. The researchers are now investigating the By Joanne Kotz, Senior Editor small molecule’s effects in other neurodegenerative and A California team has shown that monoacylglycerol lipase, which con- neurological diseases, and newco Abide Therapeutics is trols levels of a pain-reducing metabolite in the brain, also regulates exercising an option to license the MAGL inhibitors. neuroinflammation.1 The researchers have proof of concept that a small TARGETS & MECHANISMS molecule inhibitor of the target blocks inflammation and decreases 4 SEMA4D in osteoporosis neurodegeneration in a mouse model of Parkinson’s disease, and they Tokyo researchers have suggested that blocking the are now studying the compound in additional neurodegenerative and membrane protein SemA4D could directly promote bone formation in osteoporosis, and they plan to investigate the neurological diseases. target for arthritis and bone metastasis. Vaccinex already Newco Abide Therapeutics is exercising an option to license the has an inhibitor in Phase I trials for cancer. monoacylglycerol lipase (MAGL) inhibitors. MAGL is a serine hydrolase enzyme that degrades TRANSLATIONAL NOTES 2-arachidonoylglycerol (2-AG), a ligand of pain-relieving cannabinoid 6 Cancer matchmaker Cancer research UK is brokering partnerships between its receptors in the brain. In 2008, stable of academics and companies looking for a stake in a team led by Benjamin Cravatt “Targeting MAGL might early cancer discoveries. AstraZeneca is the charity’s anchor identified a brain-permeable provide a combined pain collaborator with three projects, including one involving small molecule inhibitor of and anti-inflammatory Senectus Therapeutics. -
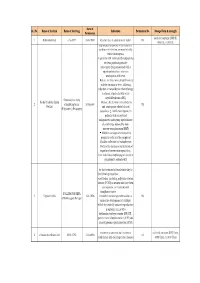
Final List of R-DNA Based Drugs Approved in the Country.Xlsx
Date of Sr. No Name of the firm Name of the Drug Indication Permission No. Dosage Form & strength Permission each vial contains 2000 IU, 1 Ethnor Limited r-hu-EPO 24-8-1993 Anaemia due to chronic renal failure Nil 4000 IU, 10,000 IU filgrastim is indicated to Decrease the incidence of infection‚ as manifested by febrile neutropenia‚ in patients with nonmyeloid malignancies receiving myelosuppressive anti-cancer drugs associated with a significant incidence of severe neutropenia with fever Reduce the time to neutrophil recovery and the duration of fever, following induction or consolidation chemotherapy treatment of patients with acute myeloid leukemia (AML) Granulocyte colony Roche Products (India) Reduce the duration of neutropenia 2 stimulating factor 30-Sep-93 Nil Pvt.Ltd and neutropenia-related clinical (Filgrastim ) (Neupogen) sequelae‚ e.g.‚ febrile neutropenia, in patients with nonmyeloid malignancies undergoing myeloablative chemotherapy followed by bone marrow transplantation (BMT) Mobilize autologous hematopoietic progenitor cells into the peripheral blood for collection by leukapheresis Reduce the incidence and duration of sequelae of severe neutropenia (e.g.‚ fever‚ infections‚ oropharyngeal ulcers) in symptomatic patients with congenital neutropenia‚ cyclic for the treatment of female infertility in the following situations: - anovulation (including polycystic ovarian disease, PCOD) in women who have been unresponsive to treatment with clomiphene citrate - FOLLITROPIN BETA 3 Organon India 14-1-1996 controlled ovarian -

Pharmacology's Review File (Endocrine)
Y Summary of (Hyperthyroidism &Hypothyroidism) *** *** *** *** *** *** *** *** *** *** *** *** *** *** *** *** Q1: Patient with which one of the following drugs is need to be assessed with the thyroid function test frequently ? A- Lithium. for Hypo B- Amiodarone. for both Hyper & Hypo. C-Both of them Q2: Which one of the following describe the main mechanism of action of propylthiouracil ? A- Block the Conversion T3 into T4 in the peripheral. B- Destroys the parenchymal cells of thyroid gland by beta rays. C- Inhibit the peroxidase enzyme centrally in thyroid gland. Q3: What is the drug of choice in treatment of hyperthyroidism in pregnant women ? A-Methimazole. B- Propylthiouracil. C- Propranolol. Q4: Why propylthiouracil is drug of choice in treatment of hyperthyroidism in pregnancy ? A- It does Crosses placenta because it hydrophobic. B- It is highly protein bound. C-It has low incidence of delayed hypothyroidism. Q5: Which one of the following is recommended to be used before thyroidectomy to decrease the possibility of bleeding from thyroid vessels ? A- Thioamides. B- Radioactive iodine. C-Lugol's solution or (KI). Q6: Which one of the following describe the mechanism of action of radioactive iodine ? A- Decrease both vascularity & size of thyroid gland by inhibiting the mitosis. B- Destroys the parenchymal cells of thyroid gland by beta rays. C- Inhibit the peroxidase enzyme centrally in thyroid gland. Q7:Which one of the following line of treatment of hyperthyroidism acting by destruction of thyroid’s parenchyma due to emission of β rays ? A- Thioamides. B- Radioactive iodine. C- Lugol's solution Q8:Which one of the following line of treatment of hyperthyroidism may safe the life of patient especially if he has tachycardia and sever palpitation a long with her hyperthyroidism ? A- Thioamides. -

Prior Authorization Medications Requiring
Prior Authorization Medications Requiring Review – Criteria for Use The Medicare Part D formulary does not allow prior authorization or criteria restrictions on medications; this document applies to the Commercial, Triple Tier, Multi-Choice, and Qualified Health Plans formularies. * The Pharmacy Consult Service reviews criteria restrictions for Diabetes, Hepatitis C, Multiple Sclerosis Medications, and PCSK9 Inhibitors Brand Name and/or Generic Name J-Code Medicare Status Notes Therapeutic Class Acthar® Corticotropin gel J0800 Medicare Part D Reviewed by the Pharmacy Consult Service for Mutiple Sclerosis indication only Aimovig™ Erenumab-aooe Medicare Part D Ampyra™ Dalfampridine Medicare Part D Reviewed by the Pharmacy Consult Service Arcalyst™ Rilonacept powder for solution J3490 Medicare Part D Aubagio® Teriflunomide Medicare Part D Reviewed by the Pharmacy Consult Service Berinert® Human C1 Inhibitor Medicare Part D Botulinum Toxin: Medicare Part D Botox® (P) Botulinum Toxins Type A J0585 (type Dysport™ (N) Botulinum Toxins Type B A) Myobloc® (N) J0587 (type Xeomin® (N) B) Cerdelga™ Eliglustat Medicare Part D Cholbam® cholic acid Medicare Part D Last Revised: August 22, 2018 CONFIDENTIAL: FOR INTERNAL KAISER PERMANENTE USE ONLY Page 1 of 91 Brand Name and/or Generic Name J-Code Medicare Status Notes Therapeutic Class Cinqair® Reslizumab Medicare Part B or D Daklinza® Daclatasvir Medicare Part D Reviewed by the Pharmacy Consult Service Dipeptidyl peptidase 4 (DPP- Medicare Part D Reviewed by the IV) inhibitors*: Pharmacy Januvia™ Sitagliptin