Dynamics of a Hepatocyte-Cholangiocyte Decision-Making Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cholangiocytes and the Environment in Primary Sclerosing Cholangitis
Gut Online First, published on July 21, 2017 as 10.1136/gutjnl-2017-314249 Leading article microbial infection (eg, Cryptosporidium Cholangiocytes and the environment in parvum)16 and PSC; (iii) accumulating evidence supporting a critical role for the Gut: first published as 10.1136/gutjnl-2017-314249 on 21 July 2017. Downloaded from primary sclerosing cholangitis: where is intestinal microbiome in the pathogenesis of PSC17; (iv) a similar genetic architecture the link? in PSC as in a prototypical exposure-driven autoimmune disease, coeliac disease18; and 1 2,3 1 (v) a similar genetic architecture in PSC as Steven P O’Hara, Tom H Karlsen, Nicholas F LaRusso 19 20 in cholestatic drug-induced injury. We began studying the cholangiopathy In primary sclerosing cholangitis (PSC), in medicine. Robert Koch in the late 19th in HIV-infected patients (a form of SSC annular fibrosis around intrahepatic and century proposed criteria to identify a due to biliary tract infection by opportu- extrahepatic bile ducts leads to progres- disease as infectious.9 These included nistic pathogens, including C parvum)21 sive liver disease for which there is no (i) the organism is regularly associated both because it was a significant clinical effective therapy except liver transplanta- with the disease, (ii) the organism can problem and as a result of our own clinical tion.1 The concentric accumulation of be isolated from the diseased host and experience with C parvum-induced SSC.22 connective tissue around bile ducts grown in culture and (iii) the disease To summarise key points from multiple suggests that the cholangiocyte plays an can be reproduced when the organism in vitro and in vivo studies,16 23 we found integral role in PSC pathogenesis. -

Expression of Specific Hepatocyte and Cholangiocyte Transcription Factors
Laboratory Investigation (2008) 88, 865–872 & 2008 USCAP, Inc All rights reserved 0023-6837/08 $30.00 Expression of specific hepatocyte and cholangiocyte transcription factors in human liver disease and embryonic development Pallavi B Limaye, Gabriela Alarco´n, Andrew L Walls, Michael A Nalesnik, George K Michalopoulos, Anthony J Demetris and Erin R Ochoa Transcription factors are major determinants of cell-specific gene expression in all cell types. Studies in rodent liver have shown that alterations in transcription factor expression determine lineage specification during fetal liver development and signify transdifferentiation of cells of the biliary compartment into ‘oval’ cells and eventually hepatocytes in adult liver. We examined the cellular localization of hepatocyte- or BEC-associated transcription factors in human fetal and adult liver and in diseases in which transdifferentiation between hepatocytes and biliary cells may play a role. In the normal adult human liver, hepatocyte nuclear factor (HNF)4a and HNF6 appeared exclusively in hepatocytes; HNF1b, HNF3a, and HNF3b were observed only in BEC. During fetal development both BEC and hepatocytes expressed HNF3a, HNF3b, and HNF6. HNF1a was expressed only in fetal hepatocytes. We further examined expression of transcription factors in massive hepatic necrosis and in specific types of chronic liver disease. Hepatocyte-associated transcription factors HNF4a and HNF6 also appeared in BEC in massive hepatic necrosis and chronic hepatitis C virus infection. Similarly, HNF3b that is expressed only in BEC in normal adult liver was also observed in hepatocytes in primary biliary cirrhosis and chronic biliary obstruction. These data mimic previous findings in rodents in which hepatocyte-associated transcription factors appear in biliary cells prior to emergence of oval cells, which function as progenitor cells for hepatocytes when the regenerative capacity of the latter is compromised. -

Liver Disease in Cystic Fibrosis
Paediatric Respiratory Reviews 15 (2014) 69–74 Contents lists available at SciVerse ScienceDirect Paediatric Respiratory Reviews Review Liver Disease in Cystic Fibrosis 1,2 1,3 3,4,5, Lisette Leeuwen , Dominic A. Fitzgerald , Kevin J. Gaskin * 1 Department of Respiratory Medicine, The Children’s Hospital at Westmead, Sydney, Australia 2 Medical School, University of Groningen, Groningen, The Netherlands 3 Discipline of Paediatrics and Child Health, Sydney Medical School, University of Sydney, Australia 4 Department of Gastroenterology, The Children’s Hospital at Westmead, Sydney, Australia 5 James Fairfax Institute of Paediatric Nutrition, University of Sydney, Sydney, Australia EDUCATIONAL AIMS To provide an overview of liver disease in children with cystic fibrosis. To review the incidence, pathogenesis, diagnosis, risk factors, outcomes and management of Cystic Fibrosis-associated Liver Disease. To describe the controversies about Cystic Fibrosis-associated Liver Disease existing in the current literature. A R T I C L E I N F O S U M M A R Y Keywords: The survival of patients with cystic fibrosis (CF) has progressively increased over recent decades, largely Cystic Fibrosis attributable to early diagnosis through newborn screening and advances in nutritional and respiratory Cystic Fibrosis-associated Liver Disease care. As the life expectancy of patients with CF has improved, non-respiratory complications such as liver Cholestasis Morbidity disease have become increasingly recognized. Mortality Biochemical derangements of liver enzymes in CF are common and may be attributed to a number of Liver transplantation specific hepatobiliary abnormalities. Among them, Cystic Fibrosis-associated Liver Disease (CFLD) is clinically the most significant hepatic complication and is believed to have a significant impact on morbidity and mortality. -
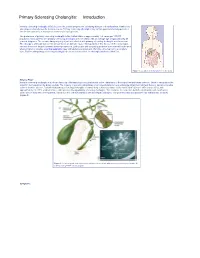
Primary Sclerosing Cholangitis: Introduction
Primary Sclerosing Cholangitis: Introduction Primary sclerosing cholangitis (PSC) is a chronic , usually progressive, stricturing disease of the biliary tree. Remissions and relapses characterize the disease course. Primary sclerosing cholangitis may remain quiescent for long periods of time in some patients; in most cases, however, it is progressive. The prevalence of primary sclerosing cholangitis in the United States is approximately 1–6 cases per 100,000 population. Most patients with primary sclerosing cholangitis are men (75%) with an average age of approximately 40 years at diagnosis. The overwhelming majority of patients affected with primary sclerosing cholangitis are Caucasian. The etiology is unknown but current opinion favors an immune cause. Management of this disease in the early stages involves the use of drugs to prevent disease progression. Endoscopic and surgical approaches are reserved for the time when symptoms develop. Liver transplantation may ultimately be required and offers the only chance for a complete cure. Patients with primary sclerosing cholangitis are at an increased risk for cholangiocarcinoma (10–15%). Figure 1. Location of the biliary tree in the body. What is PSC? Primary sclerosing cholangitis is a chronic fibrosing inflammatory process that results in the obliteration of the biliary tree and biliary cirrhosis. There is variability in the extent of involvement of the biliary system. The majority of patients with primary sclerosing cholangitis have underlying inflammatory bowel disease, namely ulcerative colitis or Crohn’s disease. Patients with primary sclerosing cholangitis are more likely to have ulcerative colitis than Crohn’s disease (85% versus 15%), with approximately 2.5–7.5% of all ulcerative colitis patients having primary sclerosing cholangitis. -

Alagille Syndrome: About Two Cases and Literature Review
Asian Journal of Pediatric Research 4(3): 1-6, 2020; Article no.AJPR.59593 ISSN: 2582-2950 Alagille Syndrome: About Two Cases and Literature Review M. Akhrif1*, A. Radi1, M. Kmari1, A. Ourrai1, A. Hassani1, R. Abilkassem1 1 and A. Agadr 1Department of Pediatric, Mohamed V Military Training Hospital, Rabat, Morocco. Authors’ contributions This work was carried out in collaboration among all authors. Authors MA and AR designed the study, performed the statistical analysis, wrote the protocol and wrote the first draft of the manuscript. Authors MA and AR managed the analyses of the study. Author MA managed the literature searches. All authors read and approved the final manuscript. Article Information DOI: 10.9734/AJPR/2020/v4i330148 Editor(s): (1) Dr. Thomaz Rafael Gollop, Universidade de Sao Paulo, Brazil. Reviewers: (1) Boris Luis Garcia Freire, Havana University, Cuba. (2) Ana Cristina Ruiz Peña, University of Zaragoza, Spain. Complete Peer review History: http://www.sdiarticle4.com/review-history/59593 Received 05 June 2020 Accepted 10 August 2020 Case Study Published 22 August 2020 ABSTRACT Alagille syndrome is a multi-systemc genetic disorder with variable phenotypic penetrance that was first described in 1969 by Daniel Alagille.It is characterized by anomalies of the intrahepatic bile ducts, heart, eye and skeleton, which are associated with facial features . The prognosis depends on the severity of the liver and heart diseases. The authors reported two cases characterized by the variability of clinical expression and evolution. The study concerned two girls aged of 2 and 4 months with no family history, who developed cholestatic jaundice evolving from the first month of life. -

Cyclic AMP Signaling in Biliary Proliferation: a Possible Target for Cholangiocarcinoma Treatment?
cells Review Cyclic AMP Signaling in Biliary Proliferation: A Possible Target for Cholangiocarcinoma Treatment? Leonardo Baiocchi 1, Ilaria Lenci 1, Martina Milana 1, Lindsey Kennedy 2,3, Keisaku Sato 2 , Wenjun Zhang 4, Burcin Ekser 4 , Ludovica Ceci 2, Vik Meadows 2, Shannon Glaser 5, Gianfranco Alpini 2,3,*,† and Heather Francis 2,3,*,† 1 Hepatology Unit, University of Tor Vergata, 00133 Rome, Italy; [email protected] (L.B.); [email protected] (I.L.); [email protected] (M.M.) 2 Hepatology and Gastroenterology, Medicine, Indiana University, Indianapolis, IN 46202, USA; [email protected] (L.K.); [email protected] (K.S.); [email protected] (L.C.); [email protected] (V.M.) 3 Richard L. Roudebush VA Medical Center, Indianapolis, IN 46202, USA 4 Division of Transplant Surgery, Department of Surgery, Indiana University, Indianapolis, IN 46202, USA; [email protected] (W.Z.); [email protected] (B.E.) 5 Department of Medical Physiology, Texas A&M University College of Medicine, Bryan, TX 77807, USA; [email protected] * Correspondence: [email protected] (G.A.); [email protected] (H.F.) † These authors contributed equally to this work. Abstract: Cholangiocarcinoma is a lethal disease with scarce response to current systemic therapy. The rare occurrence and large heterogeneity of this cancer, together with poor knowledge of its molecular mechanisms, are elements contributing to the difficulties in finding an appropriate cure. Cholangiocytes (and their cellular precursors) are considered the liver component giving rise to Citation: Baiocchi, L.; Lenci, I.; cholangiocarcinoma. These cells respond to several hormones, neuropeptides and molecular stimuli Milana, M.; Kennedy, L.; Sato, K.; employing the cAMP/PKA system for the translation of messages in the intracellular space. -

Biology of Cholangiocytes: from Bench to Bedside
Gut and Liver, Vol. 10, No. 5, September 2016, pp. 687-698 Review Biology of Cholangiocytes: From Bench to Bedside Kyo-Sang Yoo1, Woo Taek Lim2, and Ho Soon Choi1 1Department of Internal Medicine, Hanyang University College of Medicine, and 2Korea University School of Medicine, Seoul, Korea Cholangiocytes, the lining epithelial cells in bile ducts, are in human. A layer of epithelial cells called cholangiocytes lines an important subset of liver cells. They are activated by the intrahepatic bile ducts of this extensive network. The ex- endogenous and exogenous stimuli and are involved in the trahepatic ductal epithelial cells and gallbladder epithelial cells modification of bile volume and composition. They are also (GBECs) share many features with cholangiocytes. Cholangio- involved in damaging and repairing the liver. Cholangiocytes cytes comprise only about 3% to 5% of the total cell mass of the have many functions including bile production. They are also liver, but they are crucial for normal physiologic processes, and involved in transport processes that regulate the volume and they contribute to multiple disease states of the biliary tract.1-5 composition of bile. Cholangiocytes undergo proliferation Cholangiocytes serve several functions performed by several and cell death under a variety of conditions. Cholangiocytes important molecules. Most importantly, cholangiocytes partici- have functional and morphological heterogenecity. The im- pate in the formation and transportation of bile via transmem- munobiology of cholangiocytes is important, particularly for brane molecules that are expressed on the apical or basolateral understanding biliary disease. Secretion of different proin- membrane. These transporters include channels (i.e., water flammatory mediators, cytokines, and chemokines suggests channels [aquaporins]), transporters (i.e., SGLT1: Na+-glucose – – the major role that cholangiocytes play in inflammatory reac- transporter), and exchangers (i.e., SLC4A2: Cl /HCO3 exchanger). -

Characterization of Cultured Cholangiocytes Isolated from Livers
Laboratory Investigation (2014) 94, 1126–1133 & 2014 USCAP, Inc All rights reserved 0023-6837/14 Characterization of cultured cholangiocytes isolated from livers of patients with primary sclerosing cholangitis James H Tabibian1,2,4, Christy E Trussoni1,2,4, Steven P O’Hara1,2, Patrick L Splinter1,2, Julie K Heimbach3 and Nicholas F LaRusso1,2 Primary sclerosing cholangitis (PSC) is a chronic, idiopathic cholangiopathy. The role of cholangiocytes (biliary epithelial cells) in PSC pathogenesis is unknown and remains an active area of research. Here, through cellular, molecular and next- generation sequencing (NGS) methods, we characterize and identify phenotypic and signaling features of isolated PSC patient-derived cholangiocytes. We isolated cholangiocytes from stage 4 PSC patient liver explants by dissection, differential filtration and immune-magnetic bead separation. We maintained cholangiocytes in culture and assessed for: (i) cholangiocyte, cell adhesion and inflammatory markers; (ii) proliferation rate; (iii) transepithelial electrical resistance (TEER); (iv) cellular senescence; and (v) transcriptomic profiles by NGS. We used two well-established normal human cholangiocyte cell lines (H69 and NHC) as controls. Isolated PSC cells expressed cholangiocyte (eg, cytokeratin 7 and 19) and epithelial cell adhesion markers (EPCAM, ICAM) and were negative for hepatocyte and myofibroblast markers (albumin, a-actin). Proliferation rate was lower for PSC compared with normal cholangiocytes (4 vs 2 days, respectively, Po0.01). Maximum TEER was also lower in PSC compared with normal cholangiocytes (100 vs 145 Ocm2, Po0.05). Interleukin-6 (IL-6) and IL-8 (protein and mRNA) were both increased compared with NHCs and H69s (all Po0.01). The proportion of cholangiocytes staining positive for senescence-associated b-galactosidase was higher in PSC cholangio- cytes compared with NHCs (48% vs 5%, Po0.01). -

Morphological and Functional Heterogeneity of the Mouse Intrahepatic Biliary Epithelium
Laboratory Investigation (2009) 89, 456–469 & 2009 USCAP, Inc All rights reserved 0023-6837/09 $32.00 Morphological and functional heterogeneity of the mouse intrahepatic biliary epithelium Shannon S Glaser1,2,15,*, Eugenio Gaudio3,15, Arundhati Rao4,5,6, Lisa M Pierce7, Paolo Onori8, Antonio Franchitto3, Heather L Francis2,9, David E Dostal10,11, Julie K Venter2, Sharon DeMorrow1,2, Romina Mancinelli1,3, Guido Carpino12, Domenico Alvaro13, Shelley E Kopriva10, Jennifer M Savage2 and Gianfranco D Alpini1,2,10,14 Rat and human biliary epithelium is morphologically and functionally heterogeneous. As no information exists on the heterogeneity of the murine intrahepatic biliary epithelium, and with increased usage of transgenic mouse models to study liver disease pathogenesis, we sought to evaluate the morphological, secretory, and proliferative phenotypes of small and large bile ducts and purified cholangiocytes in normal and cholestatic mouse models. For morphometry, normal and bile duct ligation (BDL) mouse livers (C57/BL6) were dissected into blocks of 2–4 mm2, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Sizes of bile ducts and cholangiocytes were evaluated by using SigmaScan to measure the diameters of bile ducts and cholangiocytes. In small and large normal and BDL cholangiocytes, we evaluated the expression of cholangiocyte-specific markers, keratin-19 (KRT19), secretin receptor (SR), cystic fibrosis À À transmembrane conductance regulator (CFTR), and chloride bicarbonate anion exchanger 2 (Cl /HCO3 AE2) by im- munofluorescence and western blot; and intracellular cyclic adenosine 30,50-monophosphate (cAMP) levels and chloride efflux in response to secretin (100 nM). To evaluate cholangiocyte proliferative responses after BDL, small and large cholangiocytes were isolated from BDL mice. -

Cholangiocytes in the Pathogenesis of Primary Sclerosing Cholangitis and Development of Cholangiocarcinoma Chung, Brian; Karlsen, Tom Hemming; Folseraas, Trine
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by University of Birmingham Research Portal Cholangiocytes in the pathogenesis of primary sclerosing cholangitis and development of cholangiocarcinoma Chung, Brian; Karlsen, Tom Hemming; Folseraas, Trine DOI: 10.1016/j.bbadis.2017.08.020 License: Creative Commons: Attribution-NonCommercial-NoDerivs (CC BY-NC-ND) Document Version Peer reviewed version Citation for published version (Harvard): Chung, B, Karlsen, TH & Folseraas, T 2017, 'Cholangiocytes in the pathogenesis of primary sclerosing cholangitis and development of cholangiocarcinoma', Biochimica et Biophysica Acta. Molecular Basis of Disease. https://doi.org/10.1016/j.bbadis.2017.08.020 Link to publication on Research at Birmingham portal General rights Unless a licence is specified above, all rights (including copyright and moral rights) in this document are retained by the authors and/or the copyright holders. The express permission of the copyright holder must be obtained for any use of this material other than for purposes permitted by law. •Users may freely distribute the URL that is used to identify this publication. •Users may download and/or print one copy of the publication from the University of Birmingham research portal for the purpose of private study or non-commercial research. •User may use extracts from the document in line with the concept of ‘fair dealing’ under the Copyright, Designs and Patents Act 1988 (?) •Users may not further distribute the material nor use it for the purposes of commercial gain. Where a licence is displayed above, please note the terms and conditions of the licence govern your use of this document. -

Cholangiocytes Express the Aquaporin CHIP and Transport Water Via a Channel-Mediated Mechanism STUART K
Proc. Natl. Acad. Sci. USA Vol. 91, pp. 13009-13013, December 1994 Physiology Cholangiocytes express the aquaporin CHIP and transport water via a channel-mediated mechanism STUART K. ROBERTS*, MOTOYOSHI YANO*, YOSHIYUKI UENO*, LINH PHAM*, GIANFRANCO ALPINI*, PETER AGREt, AND NICHOLAS F. LARUSSO*t Departments of *Internal Medicine and tBiochemistry and Molecular Biology, Mayo Clinic, Rochester, MN 55905; and tDepartments of Biological Chemistry and Medicine, Johns Hopkins University, Baltimore, MD 21205 Communicated by Ralph T. Holman, August 15, 1994 (receivedfor review April 6, 1994) ABSTRACT Cholangiocytes line the intrahepatic bile pore capable of transporting water across the plasma mem- ducts and regulate salt and water secretion during bile forma- brane in a rapid, relatively temperature-independent and tion, but the mechanism(s) regulating ductal water movement mercury-sensitive manner. Moreover, immunohistochemical remains obscure. A water-selective channel, the aquaporin (13-16) and Northern blot (10, 17, 18) analyses and in situ CHIP, was recently described in several epithelia, so we tested hybridization (19) have demonstrated that CHIP has a wide the hypothesis that osmotic water movement by cholangiocytes tissue distribution, suggesting that it might be a general water is mediated by CHIP. Isolated rodent cholangiocytes showed a channel (9). Thus, we began to examine the mechanism(s) by rapid increase in volume in the presence of hypotonic extra- which water traverses biliary epithelia. cellular buffers; the ratio ofosmotic to diffusional permeability coefficients was >10. The osmotically induced increase in MATERIALS AND METHODS cholangiocyte volume was inversely proportional to buffer Cholangiocytes. Cholangiocytes [>95% pure by specific osmolality, independent of temperature, and reversibly markers (20)] were isolated from livers of male Fiseher rats blocked by HgCl2. -

Biology of Cholangiocytes: from Bench to Bedside
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by HANYANG Repository Gut and Liver, Vol. 10, No. 5, September 2016, pp. 687-698 Review Biology of Cholangiocytes: From Bench to Bedside Kyo-Sang Yoo1, Woo Taek Lim2, and Ho Soon Choi1 1Department of Internal Medicine, Hanyang University College of Medicine, and 2Korea University School of Medicine, Seoul, Korea Cholangiocytes, the lining epithelial cells in bile ducts, are in human. A layer of epithelial cells called cholangiocytes lines an important subset of liver cells. They are activated by the intrahepatic bile ducts of this extensive network. The ex- endogenous and exogenous stimuli and are involved in the trahepatic ductal epithelial cells and gallbladder epithelial cells modification of bile volume and composition. They are also (GBECs) share many features with cholangiocytes. Cholangio- involved in damaging and repairing the liver. Cholangiocytes cytes comprise only about 3% to 5% of the total cell mass of the have many functions including bile production. They are also liver, but they are crucial for normal physiologic processes, and involved in transport processes that regulate the volume and they contribute to multiple disease states of the biliary tract.1-5 composition of bile. Cholangiocytes undergo proliferation Cholangiocytes serve several functions performed by several and cell death under a variety of conditions. Cholangiocytes important molecules. Most importantly, cholangiocytes partici- have functional and morphological heterogenecity. The im- pate in the formation and transportation of bile via transmem- munobiology of cholangiocytes is important, particularly for brane molecules that are expressed on the apical or basolateral understanding biliary disease.