Blueprint Genetics Comprehensive Short Stature Syndrome Panel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
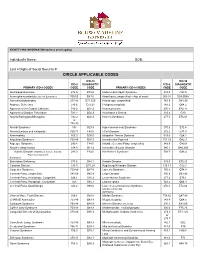
Circle Applicable Codes
IDENTIFYING INFORMATION (please print legibly) Individual’s Name: DOB: Last 4 Digits of Social Security #: CIRCLE APPLICABLE CODES ICD-10 ICD-10 ICD-9 DIAGNOSTIC ICD-9 DIAGNOSTIC PRIMARY ICD-9 CODES CODE CODE PRIMARY ICD-9 CODES CODE CODE Abetalipoproteinemia 272.5 E78.6 Hallervorden-Spatz Syndrome 333.0 G23.0 Acrocephalosyndactyly (Apert’s Syndrome) 755.55 Q87.0 Head Injury, unspecified – Age of onset: 959.01 S09.90XA Adrenaleukodystrophy 277.86 E71.529 Hemiplegia, unspecified 342.9 G81.90 Arginase Deficiency 270.6 E72.21 Holoprosencephaly 742.2 Q04.2 Agenesis of the Corpus Callosum 742.2 Q04.3 Homocystinuria 270.4 E72.11 Agenesis of Septum Pellucidum 742.2 Q04.3 Huntington’s Chorea 333.4 G10 Argyria/Pachygyria/Microgyria 742.2 Q04.3 Hurler’s Syndrome 277.5 E76.01 or 758.33 Aicardi Syndrome 333 G23.8 Hyperammonemia Syndrome 270.6 E72.4 Alcohol Embryo and Fetopathy 760.71 F84.5 I-Cell Disease 272.2 E77.0 Anencephaly 655.0 Q00.0 Idiopathic Torsion Dystonia 333.6 G24.1 Angelman Syndrome 759.89 Q93.5 Incontinentia Pigmenti 757.33 Q82.3 Asperger Syndrome 299.8 F84.5 Infantile Cerebral Palsy, unspecified 343.9 G80.9 Ataxia-Telangiectasia 334.8 G11.3 Intractable Seizure Disorder 345.1 G40.309 Autistic Disorder (Childhood Autism, Infantile 299.0 F84.0 Klinefelter’s Syndrome 758.7 Q98.4 Psychosis, Kanner’s Syndrome) Biotinidase Deficiency 277.6 D84.1 Krabbe Disease 333.0 E75.23 Canavan Disease 330.0 E75.29 Kugelberg-Welander Disease 335.11 G12.1 Carpenter Syndrome 759.89 Q87.0 Larsen’s Syndrome 755.8 Q74.8 Cerebral Palsy, unspecified 343.69 G80.9 -

Repercussions of Inborn Errors of Immunity on Growth☆ Jornal De Pediatria, Vol
Jornal de Pediatria ISSN: 0021-7557 ISSN: 1678-4782 Sociedade Brasileira de Pediatria Goudouris, Ekaterini Simões; Segundo, Gesmar Rodrigues Silva; Poli, Cecilia Repercussions of inborn errors of immunity on growth☆ Jornal de Pediatria, vol. 95, no. 1, Suppl., 2019, pp. S49-S58 Sociedade Brasileira de Pediatria DOI: https://doi.org/10.1016/j.jped.2018.11.006 Available in: https://www.redalyc.org/articulo.oa?id=399759353007 How to cite Complete issue Scientific Information System Redalyc More information about this article Network of Scientific Journals from Latin America and the Caribbean, Spain and Journal's webpage in redalyc.org Portugal Project academic non-profit, developed under the open access initiative J Pediatr (Rio J). 2019;95(S1):S49---S58 www.jped.com.br REVIEW ARTICLE ଝ Repercussions of inborn errors of immunity on growth a,b,∗ c,d e Ekaterini Simões Goudouris , Gesmar Rodrigues Silva Segundo , Cecilia Poli a Universidade Federal do Rio de Janeiro (UFRJ), Faculdade de Medicina, Departamento de Pediatria, Rio de Janeiro, RJ, Brazil b Universidade Federal do Rio de Janeiro (UFRJ), Instituto de Puericultura e Pediatria Martagão Gesteira (IPPMG), Curso de Especializac¸ão em Alergia e Imunologia Clínica, Rio de Janeiro, RJ, Brazil c Universidade Federal de Uberlândia (UFU), Faculdade de Medicina, Departamento de Pediatria, Uberlândia, MG, Brazil d Universidade Federal de Uberlândia (UFU), Hospital das Clínicas, Programa de Residência Médica em Alergia e Imunologia Pediátrica, Uberlândia, MG, Brazil e Universidad del Desarrollo, -

Dwarfism Awareness
LPA Mission Statement LPA is dedicated to improving the quality of life for people with dwarfism throughout their lives, while celebrating with great pride little people’s contribution to social diversity. LPA strives to bring solutions and global For More Information awareness to the prominent issues affecting individuals of Contact LPA short stature and their families. Toll Free…(888) LPA-2001 Direct....(714) 368-3689 Fax…..(707) 721-1896 Dwarfism Check out our website at Awareness www.lpaonline.org A Community Outreach Program 617 Broadway #518 Sponsored by Little People of America Sonoma, CA 95476 LPA is a non-profit tax exempt 501(c)3 organization funded by individual donations. Contact LPA to help. Dwarfism - Facts and Fiction Mythbusters Terminology Bodies come in all shapes and sizes. There are about 400 People with dwarfism are not magical; they do not fly, nor are Preferred terminology is a personal decision, but different types of dwarfism. Each type of dwarfism is they leprechauns, elves, fairies or any other mythological commonly accepted terms are - short stature, different than the other. Many types of dwarfism have creature. They are people - people whose bones happen to dwarfism, little person, dwarf. And we say some medical complications but most people have an grow differently than yours. That is all. "average-height" instead of "normal height". average lifespan, being productive members of society. People with dwarfism do not all know each People with dwarfism are different, yes, but not Eighty percent of people with dwarfism have average- other or look alike, nor are there towns "abnormal". height parents and siblings. -

Prevalence and Incidence of Rare Diseases: Bibliographic Data
Number 1 | January 2019 Prevalence and incidence of rare diseases: Bibliographic data Prevalence, incidence or number of published cases listed by diseases (in alphabetical order) www.orpha.net www.orphadata.org If a range of national data is available, the average is Methodology calculated to estimate the worldwide or European prevalence or incidence. When a range of data sources is available, the most Orphanet carries out a systematic survey of literature in recent data source that meets a certain number of quality order to estimate the prevalence and incidence of rare criteria is favoured (registries, meta-analyses, diseases. This study aims to collect new data regarding population-based studies, large cohorts studies). point prevalence, birth prevalence and incidence, and to update already published data according to new For congenital diseases, the prevalence is estimated, so scientific studies or other available data. that: Prevalence = birth prevalence x (patient life This data is presented in the following reports published expectancy/general population life expectancy). biannually: When only incidence data is documented, the prevalence is estimated when possible, so that : • Prevalence, incidence or number of published cases listed by diseases (in alphabetical order); Prevalence = incidence x disease mean duration. • Diseases listed by decreasing prevalence, incidence When neither prevalence nor incidence data is available, or number of published cases; which is the case for very rare diseases, the number of cases or families documented in the medical literature is Data collection provided. A number of different sources are used : Limitations of the study • Registries (RARECARE, EUROCAT, etc) ; The prevalence and incidence data presented in this report are only estimations and cannot be considered to • National/international health institutes and agencies be absolutely correct. -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Update on New GH-IGF Axis Genetic Defects
review Update on new GH-IGF axis genetic defects Gabriela A. Vasques1,2 1 Unidade de Endocrinologia https://orcid.org/0000-0002-6455-8682 Genética, Laboratório de Endocrinologia Celular e Nathalia L. M. Andrade1,2 https://orcid.org/0000-0002-1628-7881 Molecular (LIM25), Hospital das Clínicas, Faculdade de Fernanda A. Correa2 Medicina, Universidade de São https://orcid.org/0000-0003-2107-9494 Paulo, São Paulo, SP, Brasil 2 Alexander A. L. Jorge1,2 Unidade de Endocrinologia do https://orcid.org/0000-0003-2567-7360 Desenvolvimento, Laboratório de Hormônios e Genética Molecular (LIM42), Hospital das Clínicas, Faculdade de ABSTRACT Medicina, Universidade de São The somatotropic axis is the main hormonal regulator of growth. Growth hormone (GH), also known Paulo, São Paulo, SP, Brasil as somatotropin, and insulin-like growth factor 1 (IGF-1) are the key components of the somatotropic axis. This axis has been studied for a long time and the knowledge of how some molecules could Correspondence to: promote or impair hormones production and action has been growing over the last decade. The Alexander A. L. Jorge Laboratório de Endocrinologia enhancement of large-scale sequencing techniques has expanded the spectrum of known genes and Celular e Molecular (LIM25), several other candidate genes that could affect the GH-IGF1-bone pathway. To date, defects in more Faculdade de Medicina, than forty genes were associated with an impairment of the somatotropic axis. These defects can Universidade de São Paulo Av. Dr. Arnaldo, 455, affect from the secretion of GH to the bioavailability and action of IGF-1. Affected patients present a 5º andar, sala 5.340 large heterogeneous group of conditions associated with growth retardation. -

Genetic Disorder
Genetic disorder Single gene disorder Prevalence of some single gene disorders[citation needed] A single gene disorder is the result of a single mutated gene. Disorder Prevalence (approximate) There are estimated to be over 4000 human diseases caused Autosomal dominant by single gene defects. Single gene disorders can be passed Familial hypercholesterolemia 1 in 500 on to subsequent generations in several ways. Genomic Polycystic kidney disease 1 in 1250 imprinting and uniparental disomy, however, may affect Hereditary spherocytosis 1 in 5,000 inheritance patterns. The divisions between recessive [2] Marfan syndrome 1 in 4,000 and dominant types are not "hard and fast" although the [3] Huntington disease 1 in 15,000 divisions between autosomal and X-linked types are (since Autosomal recessive the latter types are distinguished purely based on 1 in 625 the chromosomal location of Sickle cell anemia the gene). For example, (African Americans) achondroplasia is typically 1 in 2,000 considered a dominant Cystic fibrosis disorder, but children with two (Caucasians) genes for achondroplasia have a severe skeletal disorder that 1 in 3,000 Tay-Sachs disease achondroplasics could be (American Jews) viewed as carriers of. Sickle- cell anemia is also considered a Phenylketonuria 1 in 12,000 recessive condition, but heterozygous carriers have Mucopolysaccharidoses 1 in 25,000 increased immunity to malaria in early childhood, which could Glycogen storage diseases 1 in 50,000 be described as a related [citation needed] dominant condition. Galactosemia -

Laron Syndrome
Laron syndrome Description Laron syndrome is a rare form of short stature that results from the body's inability to use growth hormone, a substance produced by the brain's pituitary gland that helps promote growth. Affected individuals are close to normal size at birth, but they experience slow growth from early childhood that results in very short stature. If the condition is not treated, adult males typically reach a maximum height of about 4.5 feet; adult females may be just over 4 feet tall. Other features of untreated Laron syndrome include reduced muscle strength and endurance, low blood sugar levels (hypoglycemia) in infancy, small genitals and delayed puberty, hair that is thin and fragile, and dental abnormalities. Many affected individuals have a distinctive facial appearance, including a protruding forehead, a sunken bridge of the nose (saddle nose), and a blue tint to the whites of the eyes (blue sclerae). Affected individuals have short limbs compared to the size of their torso, as well as small hands and feet. Adults with this condition tend to develop obesity. However, the signs and symptoms of Laron syndrome vary, even among affected members of the same family. Studies suggest that people with Laron syndrome have a significantly reduced risk of cancer and type 2 diabetes. Affected individuals appear to develop these common diseases much less frequently than their unaffected relatives, despite having obesity (a risk factor for both cancer and type 2 diabetes). However, people with Laron syndrome do not seem to have an increased lifespan compared with their unaffected relatives. Frequency Laron syndrome is a rare disorder. -

Genetic Short Stature ⁎ Michelle Grunauera, Alexander A.L
Growth Hormone & IGF Research xxx (xxxx) xxx–xxx Contents lists available at ScienceDirect Growth Hormone & IGF Research journal homepage: www.elsevier.com/locate/ghir Genetic short stature ⁎ Michelle Grunauera, Alexander A.L. Jorgeb, a Pediatric Intensive Care Unit, Hospital de los Valles, Escuela de Medicina, Universidad San Francisco de Quito (USFQ), Quito, Ecuador b Unidade de Endocrinologia Genetica (LIM25), Hospital das Clinicas da Faculdade de Medicina da Universidade de São Paulo (FMUSP), Sao Paulo, Brazil ARTICLE INFO ABSTRACT Keywords: Adult height and growth patterns are largely genetically programmed. Studies in twins have indicated that the Genetic heritability of height is high (> 80%), suggesting that genetic variation is the main determinant of stature. Short stature Height exhibits a normal (Gaussian) distribution according to sex, age, and ancestry. Short stature is usually Skeletal dysplasia defined as a height which is 2 standard deviations (S.D.) less than the mean height of a specific population. This Syndrome definition includes 2.3% of the population and usually includes healthy individuals. In this group of short stature Adult height non-syndromic conditions, the genetic influence occurs polygenically or oligogenically. As a rule, each common genetic variant accounts for a small effect (1 mm) on individual height variation. Recently, several studies de- monstrated that some rare variants can cause greater effect on height, without causing a syndromic condition. In more extreme cases, height SDS below 2.5 or 3 (which would comprise approximately 0.6 and 0.1% of the population, respectively) is frequently associated with syndromic conditions and are usually caused by a monogenic defect. -

Fibrochondrogenesis
Fibrochondrogenesis Description Fibrochondrogenesis is a very severe disorder of bone growth. Affected infants have a very narrow chest, which prevents the lungs from developing normally. Most infants with this condition are stillborn or die shortly after birth from respiratory failure. However, some affected individuals have lived into childhood. Fibrochondrogenesis is characterized by short stature (dwarfism) and other skeletal abnormalities. Affected individuals have shortened long bones in the arms and legs that are unusually wide at the ends (described as dumbbell-shaped). People with this condition also have a narrow chest with short, wide ribs and a round and prominent abdomen. The bones of the spine (vertebrae) are flattened (platyspondyly) and have a characteristic pinched or pear shape that is noticeable on x-rays. Other skeletal abnormalities associated with fibrochondrogenesis include abnormal curvature of the spine and underdeveloped hip (pelvic) bones. People with fibrochondrogenesis also have distinctive facial features. These include prominent eyes, low-set ears, a small mouth with a long upper lip, and a small chin ( micrognathia). Affected individuals have a relatively flat-appearing midface, particularly a small nose with a flat nasal bridge and nostrils that open to the front rather than downward (anteverted nares). Vision problems, including severe nearsightedness (high myopia) and clouding of the lens of the eye (cataract), are common in those who survive infancy. Most affected individuals also have sensorineural hearing loss, which is caused by abnormalities of the inner ear. Frequency Fibrochondrogenesis appears to be a rare disorder. About 20 affected individuals have been described in the medical literature. Causes Fibrochondrogenesis can result from mutations in the COL11A1 or COL11A2 gene. -

Blueprint Genetics Comprehensive Growth Disorders / Skeletal
Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel Test code: MA4301 Is a 374 gene panel that includes assessment of non-coding variants. This panel covers the majority of the genes listed in the Nosology 2015 (PMID: 26394607) and all genes in our Malformation category that cause growth retardation, short stature or skeletal dysplasia and is therefore a powerful diagnostic tool. It is ideal for patients suspected to have a syndromic or an isolated growth disorder or a skeletal dysplasia. About Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders This panel covers a broad spectrum of diseases associated with growth retardation, short stature or skeletal dysplasia. Many of these conditions have overlapping features which can make clinical diagnosis a challenge. Genetic diagnostics is therefore the most efficient way to subtype the diseases and enable individualized treatment and management decisions. Moreover, detection of causative mutations establishes the mode of inheritance in the family which is essential for informed genetic counseling. For additional information regarding the conditions tested on this panel, please refer to the National Organization for Rare Disorders and / or GeneReviews. Availability 4 weeks Gene Set Description Genes in the Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ACAN# Spondyloepimetaphyseal dysplasia, aggrecan type, AD/AR 20 56 Spondyloepiphyseal dysplasia, Kimberley -

Supplementary Information
Supplementary Information Structural Capacitance in Protein Evolution and Human Diseases Chen Li, Liah V T Clark, Rory Zhang, Benjamin T Porebski, Julia M. McCoey, Natalie A. Borg, Geoffrey I. Webb, Itamar Kass, Malcolm Buckle, Jiangning Song, Adrian Woolfson, and Ashley M. Buckle Supplementary tables Table S1. Disorder prediction using the human disease and polymorphisms dataseta OR DR OO OD DD DO mutations mutations 24,758 650 2,741 513 Disease 25,408 3,254 97.44% 2.56% 84.23% 15.77% 26,559 809 11,135 1,218 Non-disease 27,368 12,353 97.04% 2.96% 90.14% 9.86% ahttp://www.uniprot.org/docs/humsavar [1] (see Materials and Methdos). The numbers listed are the ones of unique mutations. ‘Unclassifiied’ mutations, according to the UniProt, were not counted. O = predicted as ordered; OR = Ordered regions D = predicted as disordered; DR = Disordered regions 1 Table S2. Mutations in long disordered regions (LDRs) of human proteins predicted to produce a DO transitiona Average # disorder # disorder # disorder # order UniProt/dbSNP Protein Mutation Disease length of predictors predictors predictorsb predictorsc LDRd in D2P2e for LDRf UHRF1-binding protein 1- A0JNW5/rs7296162 like S1147L - 4 2^ 101 6 3 A4D1E1/rs801841 Zinc finger protein 804B V1195I - 3* 2^ 37 6 1 A6NJV1/rs2272466 UPF0573 protein C2orf70 Q177L - 2* 4 34 3 1 Golgin subfamily A member A7E2F4/rs347880 8A K480N - 2* 2^ 91 N/A 2 Axonemal dynein light O14645/rs11749 intermediate polypeptide 1 A65V - 3* 3 43 N/A 2 Centrosomal protein of 290 O15078/rs374852145 kDa R2210C - 2 3 123 5 1 Fanconi