Folded DNA in Action: Hairpin Formation and Biological Functions in Prokaryotes David Bikard, Céline Loot, Zeynep Baharoglu, Didier Mazel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Epigenetic Regulator Cfp1
Article in press - uncorrected proof BioMol Concepts, Vol. 1 (2010), pp. 325–334 • Copyright ᮊ by Walter de Gruyter • Berlin • New York. DOI 10.1515/BMC.2010.031 Review The epigenetic regulator Cfp1 David G. Skalnik concept is illustrated by a variety of phenomena, including Wells Center for Pediatric Research, Section of Pediatric X-chromosome inactivation, in which one X chromosome in Hematology/Oncology, Departments of Pediatrics and each cell of a developing female blastocyst becomes irre- Biochemistry and Molecular Biology, Indiana University versibly inactivated; genomic imprinting, in which mater- School of Medicine, 1044 W. Walnut St., Indianapolis, nally and paternally derived alleles of a gene are IN 46202, USA differentially expressed; and the observation that diverse tis- sues express distinct sets of genes to permit unique func- e-mail: [email protected] tional properties, yet each (with rare exceptions) carries identical genetic information (1–4). Epigenetic information is largely encoded within chro- matin structure. A major class of epigenetic modifications is Abstract post-translational modification of histones. Dozens of dis- Numerous epigenetic modifications have been identified and tinct covalent modifications at specific amino acid residues correlated with transcriptionally active euchromatin or have been identified, including acetylation, methylation, repressed heterochromatin and many enzymes responsible phosphorylation, and sumoylation (2, 5, 6). Many of these for the addition and removal of these marks have been char- modifications are tightly correlated with either transcription- acterized. However, less is known regarding how these ally active euchromatin or transcriptionally silenced hetero- enzymes are regulated and targeted to appropriate genomic chromatin. Relatively subtle changes of covalent modifica- locations. -

A Versatile Couple with Roles in Replication and Recombination
Colloquium Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: A versatile couple with roles in replication and recombination Charles E. Jones*, Timothy C. Mueser†, Kathleen C. Dudas‡, Kenneth N. Kreuzer‡, and Nancy G. Nossal*§ *Laboratory of Molecular and Cellular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892-0830; †Department of Chemistry, University of Toledo, 2801 West Bancroft Street, Toledo, OH 43606; and ‡Department of Microbiology, Duke University Medical Center, Durham, NC 27710 Bacteriophage T4 uses two modes of replication initiation: origin- protein (gene 45) that is loaded by the complex of the gene 44 dependent replication early in infection and recombination-depen- and 62 proteins. In the presence of the T4 gene 32 single- dent replication at later times. The same relatively simple complex stranded DNA binding protein, T4 DNA polymerase, the clamp, of T4 replication proteins is responsible for both modes of DNA and the clamp loader are sufficient for slow strand displacement synthesis. Thus the mechanism for loading the T4 41 helicase must synthesis of the leading strand. The 5Ј to 3Ј gene 41 helicase be versatile enough to allow it to be loaded on R loops created by unwinds DNA ahead of the fork and increases the elongation transcription at several origins, on D loops created by recombina- rate more than 10-fold to 400 nt͞sec, comparable to that in vivo. tion, and on stalled replication forks. T4 59 helicase-loading protein Although the helicase can load on nicked and forked DNA by is a small, basic, almost completely ␣-helical protein whose N- itself, its loading is greatly accelerated by the 59 helicase-loading terminal domain has structural similarity to high mobility group protein. -

Teacher Notes Science Nspired
DNA Replication TEACHER NOTES SCIENCE NSPIRED Science Objectives In this lesson, students will • Explore how the structure of DNA supports its semi-conservative replication • Identify the name and function of several key enzymes in DNA replication • Recognize the function of Okazaki fragments in DNA replication Vocabulary TI-Nspire™ Technology Skills: • Double helix • Leading strand • Download a TI-Nspire • Lagging strand • Polymerase I • Polymerase III • Helicase document • Base pairs • Okazaki Fragments • Open a document • Primase • Move between pages • Open Directions Box About the Lesson • Explore ‘Hot Spots’ • Using three simulations, students will interact with DNA replication to explore semi-conservative replication and identify Tech Tips: specific enzymes and their roles in replication. Assessments are Make sure that students know embedded in the activity to engage discussion and gauge how to move between pages by learning. pressing /¡ (left arrow) and • As a result, students will: /¢ (right arrow). • Learn the basic functions of the following DNA replication enzymes: helicase, primase, ligase, polymerase I and III. Lesson Materials: • Learn how the double helix reproduces in a semi- Student Activity • DNA_Replication_Student.do conservative fashion c • Learn the role and function of Okazaki fragments in • DNA_Replication_Student.pd replication of the lagging strand. f TI-Nspire document TI-Nspire™ Navigator™ • DNA_Replication.tns Not required. If using Navigator: • Send out the DNA_Replication.tns file. • Monitor student progress -
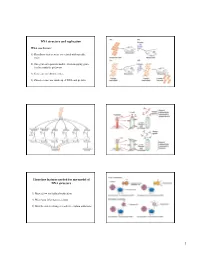
DNA Structure and Replication Three Key Features Needed for Any Model
DNA structure and replication What was known? 1) Hereditary factors were associated with specific traits 2) One-gene-one-protein model - from mapping genes for biosynthetic pathways 3) Genes are on chromosomes 4) Chromosomes are made up of DNA and protein Three key features needed for any model of DNA structure 1) Must allow for faithful replication 2) Must have information content 3) Must be able to change in order to explain mutations 1 Rosalind Franklin James Watson Francis Crick X-ray diffraction Maurice Wilkins Photograph of B-DNA Linus Pauling Nature 171, 737-738 (1953) © Macmillan Publishers Ltd. Molecular structure of Nucleic Acids WATSON, J. D. & CRICK, F. H. C. Features of the double helix A Structure for Deoxyribose Nucleic Acid 1) Two parallel strands We wish to suggest a structure for the salt of deoxyribose nucleic acid (D.N.A.). This structure has novel features which are of considerable biological interest………… 2) Bases held together by H-bonds 3) Phosphodiester backbone 4) The base is attached to the position 1 on the sugar 5) Base pair stack - provides Figure 1. This figure is purely diagrammatic. The two ribbons symbolize the two phophate-sugar chains, and the horizonal rods stability the pairs of bases holding the chains together. The vertical line marks the fibre axis. 6) Contains a major and …………….It has not escaped our notice that the specific pairing we minor groove have postulated immediately suggests a possible copying mechanism for the genetic material. Three key features needed for any model of DNA structure -

Palindromes in DNA—A Risk for Genome Stability and Implications in Cancer
International Journal of Molecular Sciences Review Palindromes in DNA—A Risk for Genome Stability and Implications in Cancer Marina Svetec Mikleni´cand Ivan Krešimir Svetec * Faculty of Food Technology and Biotechnology, University of Zagreb, Pierottijeva 6, 10000 Zagreb, Croatia; [email protected] * Correspondence: [email protected]; Tel.: +385-1483-6016 Abstract: A palindrome in DNA consists of two closely spaced or adjacent inverted repeats. Certain palindromes have important biological functions as parts of various cis-acting elements and protein binding sites. However, many palindromes are known as fragile sites in the genome, sites prone to chromosome breakage which can lead to various genetic rearrangements or even cell death. The ability of certain palindromes to initiate genetic recombination lies in their ability to form secondary structures in DNA which can cause replication stalling and double-strand breaks. Given their recombinogenic nature, it is not surprising that palindromes in the human genome are involved in genetic rearrangements in cancer cells as well as other known recurrent translocations and deletions associated with certain syndromes in humans. Here, we bring an overview of current understanding and knowledge on molecular mechanisms of palindrome recombinogenicity and discuss possible implications of DNA palindromes in carcinogenesis. Furthermore, we overview the data on known palindromic sequences in the human genome and efforts to estimate their number and distribution, as well as underlying mechanisms of genetic rearrangements specific palindromic sequences cause. Keywords: DNA palindromes; quasipalindromes; palindromic amplification; palindrome-mediated genetic recombination; carcinogenesis Citation: Svetec Mikleni´c,M.; Svetec, I.K. Palindromes in DNA—A Risk for Genome Stability and Implications in Cancer. -

DNA Topology Influences P53 Sequence-Specific DNA Binding
DNA topology influences p53 sequence-specific DNA binding through structural transitions within the target sites Eva Brázdová Jagelská, Václav Brázda, Petr Pečinka, Emil Paleček, Miroslav Fojta To cite this version: Eva Brázdová Jagelská, Václav Brázda, Petr Pečinka, Emil Paleček, Miroslav Fojta. DNA topology influences p53 sequence-specific DNA binding through structural transitions within the target sites. Biochemical Journal, Portland Press, 2008, 412 (1), pp.57-63. 10.1042/BJ20071648. hal-00478935 HAL Id: hal-00478935 https://hal.archives-ouvertes.fr/hal-00478935 Submitted on 30 Apr 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Biochemical Journal Immediate Publication. Published on 14 Feb 2008 as manuscript BJ20071648 DNA TOPOLOGY INFLUENCES P53 SEQUENCE-SPECIFIC DNA BINDING THROUGH STRUCTURAL TRANSITIONS WITHIN THE TARGET SITES Eva Brázdová Jagelskáa, Václav Brázdaa*, Petr Pečinkab, Emil Palečeka and Miroslav Fojtaa aInstitute of Biophysics, Academy of Sciences of the Czech Republic, 612 65 Brno, Czech Republic bFaculty of Science, University of Ostrava, 701 03 Ostrava, Czech Republic *Corresponding author Tel.: 420 541517231 Fax.: 420 541211293 e-mail: [email protected] Abbreviations: scDNA, supercoiled DNA; lin, linear DNA; o, oligodeoxynucleotide; SK, plasmid pBluescript SK-; fl, full length; wt, wild-type; Keywords: p53 protein / DNA binding / protein-DNA complex Short title: DNA topology affects p53 binding THIS IS NOT THE FINAL VERSION - see doi:10.1042/BJ20071648 Stage 2(a) POST-PRINT Page 1 Licenced copy. -

Processivity of DNA Polymerases: Two Mechanisms, One Goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2
Minireview 121 Processivity of DNA polymerases: two mechanisms, one goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2 Replicative DNA polymerases are highly processive Processive DNA synthesis by cellular replicases and the enzymes that polymerize thousands of nucleotides without bacteriophage T4 replicase dissociating from the DNA template. The recently Until recently, the only mechanism for high processivity determined structure of the Escherichia coli bacteriophage that was understood in detail was that utilized by cellular T7 DNA polymerase suggests a unique mechanism that replicases and the replicase of bacteriophage T4. This underlies processivity, and this mechanism may generalize mechanism involves a ring-shaped protein called a ‘DNA to other replicative polymerases. sliding clamp’ that encircles the DNA and tethers the polymerase catalytic unit to the DNA [3,4]. The three- Addresses: 1Department of Molecular Biology, Memorial Sloan- dimensional structures of several sliding clamps have been Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, 2 determined: the eukaryotic proliferating cell nuclear USA and Laboratory of DNA Replication, Howard Hughes Medical β Institute, The Rockefeller University, 1230 York Avenue, New York, NY antigen (PCNA) [5,6]; the subunit of the prokaryotic 10021, USA. DNA polymerase III [7]; and the bacteriophage T4 gene 45 protein (gp45) (J Kuriyan, personal communication) *Corresponding author. (Figure 1). The overall structure of these clamps is very E-mail: [email protected] similar; the PCNA, β subunit and gp45 rings are super- Structure 15 February 1998, 6:121–125 imposable [8]. Each ring has similar dimensions and a http://biomednet.com/elecref/0969212600600121 central cavity large enough to accommodate duplex DNA (Figure 1). -

Cruciform Structures Are a Common DNA Feature Important for Regulating Biological Processes Václav Brázda1*, Rob C Laister2, Eva B Jagelská1 and Cheryl Arrowsmith3
Brázda et al. BMC Molecular Biology 2011, 12:33 http://www.biomedcentral.com/1471-2199/12/33 REVIEW Open Access Cruciform structures are a common DNA feature important for regulating biological processes Václav Brázda1*, Rob C Laister2, Eva B Jagelská1 and Cheryl Arrowsmith3 Abstract DNA cruciforms play an important role in the regulation of natural processes involving DNA. These structures are formed by inverted repeats, and their stability is enhanced by DNA supercoiling. Cruciform structures are fundamentally important for a wide range of biological processes, including replication, regulation of gene expression, nucleosome structure and recombination. They also have been implicated in the evolution and development of diseases including cancer, Werner’s syndrome and others. Cruciform structures are targets for many architectural and regulatory proteins, such as histones H1 and H5, topoisomerase IIb, HMG proteins, HU, p53, the proto-oncogene protein DEK and others. A number of DNA-binding proteins, such as the HMGB-box family members, Rad54, BRCA1 protein, as well as PARP-1 polymerase, possess weak sequence specific DNA binding yet bind preferentially to cruciform structures. Some of these proteins are, in fact, capable of inducing the formation of cruciform structures upon DNA binding. In this article, we review the protein families that are involved in interacting with and regulating cruciform structures, including (a) the junction- resolving enzymes, (b) DNA repair proteins and transcription factors, (c) proteins involved in replication and (d) chromatin-associated proteins. The prevalence of cruciform structures and their roles in protein interactions, epigenetic regulation and the maintenance of cell homeostasis are also discussed. Keywords: cruciform structure, inverted repeat, protein-DNA binding Review representation of inverted repeats, which occurs nonran- Genome sequencing projects have inundated us with domly in the DNA of all organisms, has been noted in information regarding the genetic basis of life. -

Chromosomal Instability Mediated by Non-B DNA: Cruciform Conformation and Not DNA Sequence Is Responsible for Recurrent Translocation in Humans
Downloaded from genome.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press Letter Chromosomal instability mediated by non-B DNA: Cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans Hidehito Inagaki,1 Tamae Ohye,1 Hiroshi Kogo,1 Takema Kato,1,2 Hasbaira Bolor,2 Mariko Taniguchi,1,4 Tamim H. Shaikh,3 Beverly S. Emanuel,3 and Hiroki Kurahashi1,5 1Division of Molecular Genetics, Institute for Comprehensive Medical Science, Fujita Health University, Toyoake, Aichi 470-1192, Japan; 221st Century COE Program, Development Center for Targeted and Invasive Diagnosis and Treatment, Fujita Health University, Toyoake, Aichi 470-1192, Japan; 3Division of Human Genetics, The Children’s Hospital of Philadelphia and Department of Pediatrics, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104, USA Chromosomal aberrations have been thought to be random events. However, recent findings introduce a new paradigm in which certain DNA segments have the potential to adopt unusual conformations that lead to genomic instability and nonrandom chromosomal rearrangement. One of the best-studied examples is the palindromic AT-rich repeat (PATRR), which induces recurrent constitutional translocations in humans. Here, we established a plasmid-based model that pro- motes frequent intermolecular rearrangements between two PATRRs in HEK293 cells. In this model system, the pro- portion of PATRR plasmid that extrudes a cruciform structure correlates to the levels of rearrangement. Our data suggest that PATRR-mediated translocations are attributable to unusual DNA conformations that confer a common pathway for chromosomal rearrangements in humans. [Supplemental material is available online at www.genome.org.] Chromosomal aberrations, including translocations or deletions, (Kurahashi et al. -

Primer Release Is the Rate-Limiting Event in Lagging-Strand Synthesis
Primer release is the rate-limiting event in lagging- strand synthesis mediated by the T7 replisome Alfredo J. Hernandeza, Seung-Joo Leea, and Charles C. Richardsona,1 aDepartment of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115 Contributed by Charles C. Richardson, April 18, 2016 (sent for review December 30, 2015; reviewed by Nicholas E. Dixon and I. Robert Lehman) DNA replication occurs semidiscontinuously due to the antiparallel but also for enabling the use of short oligoribonucleotides by T7 DNA strands and polarity of enzymatic DNA synthesis. Although DNA polymerase. Critically, the primase domain also fulfills two the leading strand is synthesized continuously, the lagging strand additional roles apart from primer synthesis: it prevents disso- is synthesized in small segments designated Okazaki fragments. ciation of the extremely short tetramer, stabilizing it with the Lagging-strand synthesis is a complex event requiring repeated template, and it secures it in the polymerase active site (10, 12). cycles of RNA primer synthesis, transfer to the lagging-strand Here we show that the rate-limiting step in initiation of Okazaki polymerase, and extension effected by cooperation between DNA fragments by the T7 replisome is primer release from the primase primase and the lagging-strand polymerase. We examined events domain of gp4. In the absence of gp2.5, an additional step, distinct controlling Okazaki fragment initiation using the bacteriophage from primer release, also limits primer extension. The presence of T7 replication system. Primer utilization by T7 DNA polymerase is gp2.5 promotes efficient primer formation and primer utilization. slower than primer formation. -

Junction Ribonuclease: an Activity in Okazaki Fragment Processing
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 2244–2249, March 1998 Biochemistry Junction ribonuclease: An activity in Okazaki fragment processing RICHARD S. MURANTE,LEIGH A. HENRICKSEN, AND ROBERT A. BAMBARA* Department of Biochemistry and Biophysics, and Cancer Center, Box 712, University of Rochester School of Medicine and Dentistry, 601 Elmwood Avenue, Rochester, NY 14642 Communicated by I. Robert Lehman, Stanford University School of Medicine, Stanford, CA, December 29, 1997 (received for review September 15, 1997) ABSTRACT The initiator RNAs of mammalian Okazaki and removal of RNA primers. Reconstitution assays with both fragments are thought to be removed by RNase HI and the mouse cell and SV40 replication proteins require RNase HI for 5*-3* flap endonuclease (FEN1). Earlier evidence indicated the maturation of lagging strands in vitro (12, 13). that the cleavage site of RNase HI is 5* of the last ribonucle- Although cleavage of long RNAyDNA hybrids is random, otide at the RNA-DNA junction on an Okazaki substrate. In cleavage of certain substrates is clearly structure specific. Eder current work, highly purified calf RNase HI makes this exact et al. (16) found that when a series of ribonucleotides were cleavage in Okazaki fragments containing mismatches that followed by a 39 DNA segment and annealed to DNA to form distort the hybrid structure of the heteroduplex. Furthermore, a duplex, human RNase HI preferentially cleaved to leave a even fully unannealed Okazaki fragments were cleaved. DNA segment with a 59 monoribonucleotide. In reactions Clearly, the enzyme recognizes the transition from RNA to reconstituting Okazaki fragment processing, RNase HI from DNA on a single-stranded substrate and not the RNAyDNA calf thymus similarly cleaved the RNA-DNA strand 59 to the heteroduplex structure. -

Recognition of Local DNA Structures by P53 Protein
International Journal of Molecular Sciences Review Recognition of Local DNA Structures by p53 Protein Václav Brázda * and Jan Coufal Institute of Biophysics, Academy of Sciences of the Czech Republic v.v.i., Královopolská 135, 612 65 Brno, Czech Republic; [email protected] * Correspondence: [email protected]; Tel.: +420-541-517-231 Academic Editor: Tomoo Iwakuma Received: 24 November 2016; Accepted: 3 February 2017; Published: 10 February 2017 Abstract: p53 plays critical roles in regulating cell cycle, apoptosis, senescence and metabolism and is commonly mutated in human cancer. These roles are achieved by interaction with other proteins, but particularly by interaction with DNA. As a transcription factor, p53 is well known to bind consensus target sequences in linear B-DNA. Recent findings indicate that p53 binds with higher affinity to target sequences that form cruciform DNA structure. Moreover, p53 binds very tightly to non-B DNA structures and local DNA structures are increasingly recognized to influence the activity of wild-type and mutant p53. Apart from cruciform structures, p53 binds to quadruplex DNA, triplex DNA, DNA loops, bulged DNA and hemicatenane DNA. In this review, we describe local DNA structures and summarize information about interactions of p53 with these structural DNA motifs. These recent data provide important insights into the complexity of the p53 pathway and the functional consequences of wild-type and mutant p53 activation in normal and tumor cells. Keywords: p53 protein; local DNA structures; protein-DNA interactions 1. Introduction p53 is one of the most intensively studied tumor suppressor proteins and its regulation and relation to cancer has been reviewed extensively [1–3].