Cytoplasmic Domain of CD44 Acts As a Nuclear Transcription Regulator
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of a Heterozygous ACAN Mutation in a 15-Year-Old Boy With
Case report https://doi.org/10.6065/apem.1938198.099 Ann Pediatr Endocrinol Metab 2020;25:272-276 Identification of a heterozygous ACAN mutation in a 15-year-old boy with short stature who presented with advanced bone age: a case report and review of the literature Tae Youp Kim, MD1, Longitudinal bone growth is primarily mediated by the growth plate, which is Kyung Mi Jang, MD, PhD1, a specialized cartilaginous structure. Aggrecan, encoded by ACAN, is a primary Chang Won Keum, PhD2, proteoglycan component of the extracellular matrix in both the growth plate and Seung Hwan Oh, MD, PhD3, articular cartilage. Aggrecanopathies have emerged as a phenotype of genetic Woo Yeong Chung, MD, PhD4 skeletal disease in humans. A heterozygous ACAN mutation causes short stature, premature growth cessation, and accelerated bone age maturation. We report 1Department of Pediatrics, Yeungnam the case of a 15-year-old boy with familial short stature, with height of 149 cm University Hospital, Yeungnam Univer- (Korean standard deviation score [SDS] of -3.6) and weight of 50.5 kg (-1.48 SDS). sity College of Medicine, Daegu, Korea He presented with mild midfacial hypoplasia, frontal bossing, a broad chest, and 2Rare Genetic Disease Research Center, a short neck. The father's and mother's heights were 150 cm (-4.8 SDS) and 153 3Billion INC, Seoul, Korea cm (-1.69 SDS), respectively. The patient's bone age was 2–3 years more advanced 3 Departments of Laboratory Medicine than his chronological age, and no endocrine abnormalities were detected. Whole- 4 and Pediatrics, Inje University Busan exome sequencing followed by Sanger sequencing revealed a heterozygous ACAN Paik Hospital, Inje University College of mutation, c.512C>T (p.Ala171Val), in both the proband and his father. -

Structural Characterisation of Aggrecan in Cartilaginous Tissues and Tissue Engineered Constructs a Thesis Submitted to the Univ
Structural Characterisation of Aggrecan in Cartilaginous Tissues and Tissue Engineered Constructs A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy in the Faculty of Biology, Medicine and Health 2017 Russell J Craddock The School of Biological Sciences Division of Cell Matrix Biology and Regenerative Medicine List of Contents List of Contents ................................................................................................................. 2 List of Figures ................................................................................................................... 6 List of Tables ..................................................................................................................... 8 List of Equations ............................................................................................................... 8 List of Conference Presentations and Publications ....................................................... 8 List of Abbreviations ........................................................................................................ 9 Abstract ........................................................................................................................... 10 Declaration ...................................................................................................................... 11 Copyright Statement ...................................................................................................... 11 Acknowledgements ........................................................................................................ -

Binding of Hyaluronan and Its Neutral Analog by TSG-6 Link Domain
Binding of hyaluronan and its neutral analog by TSG-6 Link domain Roman Witasek 1, Eva Kutálková 1, Josef Hrnčiřík 1 and Marek Ingr 1,2 1Tomas Bata University in Zlín, Faculty of Technology, Department of Physics and Materials Engineering, Nám. T.G. Masaryka 5555, 76001 Zlín, Czech Republic 2Charles University, Faculty of Science, Department of Biochemistry, Hlavova 8/2030, 12843 Praha 2, Czech Republic 2nd Users’ Conference of Zlín IT4Innovations −> −> Hyaluronic acid (hyaluronan) – HA [4)-β-D-Glc pA-(1 3)-β-D-Glc pNAc-(1 ]n 1→3 1→4 1→3 1→4 1→4 • Essential component of proteoglycan - the main constituent of the extracellular matrix of connective tissues • Signaling molecule involved in carcinogenesis, inflammation and wound healing • Biological activity depends on the chain length (from MDa to short oligosaccharides) http://medinfo.ufl.edu/pa/chuck/summer/handouts/images/gag.jpg Hyaladherins – hyaluronan binding proteins TSG-6 Containing Link domain Consists of Link and CUB domain • Proteoglycan forming Link binds hyaluronan and other GAGs Aggrecan positively charged Brevican • CUB Neurocan not well known function Versican negatively charged Membrane receptors CUB CD44 – the main transmembrane receptor Link LYVE-1 – lymphatic endothelial cells Soluble receptor TSG-6 Without Link domain RHAMM – membrane receptor, partially disordered Inter-α-inhibitor Interactions of TSG-6 with glycosaminoglycans (GAGs) Several interactions of GAG oligosaccharides were identified by NMR experiments Computational details: Hyaluronan 1: K11, Y12, H45, V57, Y59, P60, I61, K63, F70, I76, Y78, R81, W88 • MD simulations carried out 2 st in GROMACS v. 5.1.1 Heparin : K34, K54, R56 + 1 mode: K20, K41, R84 ; • nd CHARMM 36 force field 2 mode: K72 • TIP3P model of water Chondroitin sulfate 3: 1st mode similar to HA ; • Water box 8 ×8×8 nm 2nd mode: K20, K34, K41, K54 • HA (GlcHA) oligosaccharides of 12 Questions: monosacharide units • TSG-6 Link domain – PDB code: 2N40 1. -

Tnfα-Stimulated Gene-6 (TSG6) Activates Macrophage PNAS PLUS Phenotype Transition to Prevent Inflammatory Lung Injury
TNFα-stimulated gene-6 (TSG6) activates macrophage PNAS PLUS phenotype transition to prevent inflammatory lung injury Manish Mittala, Chinnaswamy Tiruppathia, Saroj Nepala, You-Yang Zhaoa, Dagmara Grzycha, Dheeraj Sonia, Darwin J. Prockopb,1, and Asrar B. Malika,1 aDepartment of Pharmacology and Center for Lung and Vascular Biology, University of Illinois College of Medicine, Chicago, IL 60612 and bInstitute for Regenerative Medicine, Texas A&M University Health Science Center and College of Medicine, Bryan, TX 77807 Contributed by Darwin J. Prockop, November 3, 2016 (sent for review September 7, 2016; reviewed by Arshad Rehman and Sarah Yuan) TNFα-stimulated gene-6 (TSG6), a 30-kDa protein generated by acti- macrophages and other inflammatory cells sequestered in lungs (15). vated macrophages, modulates inflammation; however, its mechanism Macrophages in ALI are activated through the sensing of pathogen- of action and role in the activation of macrophages are not fully un- associated molecular patterns (PAMPs) and damage-associated derstood. Here we observed markedly augmented LPS-induced inflam- molecular patterns (DAMPs) via Toll-like receptors (TLRs). On −/− matory lung injury and mortality in TSG6 mice compared with WT binding of LPS to TLR4, macrophages produce proinflammatory +/+ (TSG6 ) mice. Treatment of mice with intratracheal instillation of cytokines, such as IFNγ,TNFα,andIL-1β,whichsignalthere- TSG6 prevented LPS-induced lung injury and neutrophil sequestration, cruitmentofneutrophilsandlymphocytes at the site of infection -

Ligand Binding and Signaling of HARE/Stabilin-2
biomolecules Review Ligand Binding and Signaling of HARE/Stabilin-2 Edward N. Harris * and Fatima Cabral Department of Biochemistry, University of Nebraska, Lincoln, NE 68588, USA * Correspondence: [email protected]; Tel.: +1-(402)-472-7468 Received: 18 June 2019; Accepted: 7 July 2019; Published: 11 July 2019 Abstract: The Stabilin receptors are a two-member family in the type H class of scavenger receptors. These dynamic receptors bind and internalize multiple ligands from the cell surface for the purpose of clearing extracellular material including some synthetic drugs and for sensing the external environment of the cell. Stabilin-1 was the first receptor to be cloned, though the biological activity of Hyaluronic Acid Receptor for Endocytosis (HARE)/Stabilin-2 was observed about 10 years prior to the cloning of Stabilin-1. Stabilin-1 has a more diverse expression profile among the tissues than HARE/Stabilin-2. This review will focus on HARE/Stabilin-2 and its interactions with hyaluronan, heparin, and phosphorothioate antisense oligonucleotides and what is known about how this receptor participates in signaling upon ligand binding. Keywords: stabilin; HARE; receptor-mediated endocytosis; scavenger receptor; hyaluronan 1. Initial Discovery The discovery of the cloned Stabilin receptors began with Stabilin-1 in the early 1990s. A monoclonal antibody raised against whole human spleen homogenate produced a monoclonal antibody (MS-1) against an antigen that was initially identified as a large protein that is preferentially expressed in the sinusoidal endothelia of spleen, lymph node, liver, and adrenal cortex [1]. The identified MS-1 antigen is an inducible protein not found in normal skin lesions, but rather in diseased skin and some cancers such as psoriasis, melanoma, and T-cell lymphomas. -

CXCL8 Direct Interaction with the Chemokine TSG-6 Inhibits
TSG-6 Inhibits Neutrophil Migration via Direct Interaction with the Chemokine CXCL8 This information is current as Douglas P. Dyer, Jennifer M. Thomson, Aurelie Hermant, of September 23, 2021. Thomas A. Jowitt, Tracy M. Handel, Amanda E. I. Proudfoot, Anthony J. Day and Caroline M. Milner J Immunol 2014; 192:2177-2185; Prepublished online 5 February 2014; doi: 10.4049/jimmunol.1300194 Downloaded from http://www.jimmunol.org/content/192/5/2177 Supplementary http://www.jimmunol.org/content/suppl/2014/02/05/jimmunol.130019 Material 4.DCSupplemental http://www.jimmunol.org/ References This article cites 56 articles, 27 of which you can access for free at: http://www.jimmunol.org/content/192/5/2177.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 23, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2014 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology TSG-6 Inhibits Neutrophil Migration via Direct Interaction with the Chemokine CXCL8 Douglas P. -

Domain Shuffling and the Evolution of Vertebrates
Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Domain shuffling and the evolution of vertebrates Takeshi Kawashima1, 2, 3, Shuichi Kawashima4, Chisaki Tanaka5, Miho Murai6, Masahiko Yoneda6, Nicholas H. Putnam2, Daniel S. Rokhsar2, Minoru Kanehisa4,7, Nori Satoh1, 8, Hiroshi Wada5 1: Okinawa Institute of Science and Technology, Uruma, Okinawa, 904-2234, Japan 2: Department of Energy Joint Genome Institute, Walnut Creek, CA 94598, USA 3: Japanese Society for Promotion of sciences, 6 Ichibancho, Chiyoda-ku, Tokyo, 102-8471, Japan 4: Human Genome Center, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokane-dai Minato-ku Tokyo 108-8639, Japan 5: Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba 305-8572 JAPAN 6: Aichi Prefectural College of Nursing and Health, Miriyama-ku Nagoya 463-8502 JAPAN 7: Bioinformatics Center, Institute of Chemical Research, Kyoto University, Kyoto 611-0011, Japan 8: Department of Zoology, Graduate School of Sciences, Kyoto University, Kyoto 606-8502, JAPAN Address for correspondense: Hiroshi Wada Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba 305-8572 JAPAN E-mail: [email protected], Tel & Fax: +81-29-853-4671 Takeshi Kawashima Okinawa Institute of Science and Technology, Uruma, Okinawa, 904-2234, Japan E-mail: [email protected], Tel; +1-98-921-2245, Fax; +1-98-934-5622 Running title: Domain shuffling in vertebrate genomes Key words: domain shuffling, exon shuffling, vertebrate, cartilage, chordate, notochord, ascidian, amphioxus 1 Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract The evolution of vertebrates included a number of important events: the development of cartilage, the immune system, and complicated craniofacial structures. -

Downloaded from Generate New ECM Proteins, Particularly in the Vertebrate Membranes, Interstitial Matrices, Tendons, Cartilage, Bones, and Teeth
The evolution of metazoan extracellular matrix The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Hynes, R. O. “Evolution: The Evolution of Metazoan Extracellular Matrix.” The Journal of Cell Biology 196.6 (2012): 671–679. Web. 4 May 2012. As Published http://dx.doi.org/10.1083/jcb.201109041 Publisher Rockefeller University Press, The Version Final published version Citable link http://hdl.handle.net/1721.1/70517 Terms of Use Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported Detailed Terms http://creativecommons.org/licenses/by-nc-sa/3.0/ Published March 19, 2012 JCB: Review Evolution The evolution of metazoan extracellular matrix Richard O. Hynes Howard Hughes Medical Institute, Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139 The modular domain structure of extracellular matrix (ECM) our current understanding of the diversity and evolution of ECM proteins and their genes has allowed extensive exon/ proteins and attempt to relate them to the evolution of multi- cellularity and the subsequent evolution of metazoa. domain shuffling during evolution to generate hundreds of ECM proteins. Many of these arose early during metazoan Major characteristics and categories evolution and have been highly conserved ever since. Others of ECM proteins in metazoa have undergone duplication and divergence during evolu- ECMs are, by definition, relatively or completely insoluble assemblies of proteins that form structures such as basement tion, and novel combinations of domains have evolved to Downloaded from generate new ECM proteins, particularly in the vertebrate membranes, interstitial matrices, tendons, cartilage, bones, and teeth. -
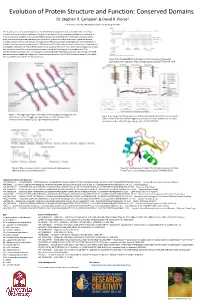
Evolution of Protein Structure and Function: Conserved Domains Dr
Evolution of Protein Structure and Function: Conserved Domains Dr. Stephen R. Campion1 & David B. Pierce1 1. Alvernia University, 400 Saint Bernardine St. Reading, PA 19607 This study is part of an ongoing exploration into the evolution of protein structure and function. Individual proteins that share a common element or domain of protein structure have been compiled and compared in order to characterize which of the available chemical functionalities inherent in the twenty common amino acids, are most heavily preserved throughout evolution, as proteins diverge over time to generate diverse proteins functions from a common primordial ancestor. This study focuses specifically on a compiled family of protein structures containing one or more LINK domains within their overall protein architecture. These domains are unique to extracellular matrix (ECM) proteins that comprise the “core” of a variety of proteoglycan structures that are critical to cellular organization and communication within biological tissues and organs. The conservation of both polar and non-polar amino acids have been identified at strategic locations with the LINK domain consensus sequence. Mapping of these amino acids within the LNK 3D structure is currently limited by the unavailability of diverse LINK structural data. Figure 3: An image highlighting the regions and interactions of GAGs and saccharides within aggrecan. https://images.app.goo.gl/GJtpT72VGVv2FHvD8 Figure 1: An image depicting the interactions between glycosaminoglycans, core proteins such as that of aggrecan, link proteins, and the hyaluronic acid Figure 4: An image illustrating a more in-depth look at chondroitin sulfate and chondroitin backbone. https://images.app.goo.gl/14GVjtHriMob2Exv5 sulfate disaccharides in relation to Aggrecan, Versican, Neurocan, and Brevican and their respecive domains. -

UCLA Electronic Theses and Dissertations
UCLA UCLA Electronic Theses and Dissertations Title Chromatin Architectural Dynamics in Cardiovascular Disease Permalink https://escholarship.org/uc/item/7wf4v4kp Author Chapski, Douglas Publication Date 2019 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA Los Angeles Chromatin Architectural Dynamics in Cardiovascular Disease A dissertation suBmitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Molecular, Cellular, and Integrative Physiology by Douglas Chapski 2019 © Copyright By Douglas Chapski 2019 ABSTRACT OF THE DISSERTATION Chromatin Architectural Dynamics in Cardiovascular Disease by Douglas Chapski Doctor of Philosophy in Molecular, Cellular, and Integrative Physiology University of California, Los Angeles, 2019 Professor Thomas M. Vondriska, Chair The chromatin architectural rearrangements that permit disease gene expression are just Beginning to come to light. Distinct levels of chromatin organization are needed to maintain a healthy transcriptome, from the histone octamer that forms nucleosomes (the functional unit of chromatin) to chromosome territories that demarcate large swaths of the nucleus. An integrative picture of hoW each level of chromatin contriButes toWards healthy and disease gene expression has eluded us until chromosome conformation capture folloWed By high-throughput sequencing paved the Way for deeper study of hoW chromatin features, such as significant chromosomal interactions, topologically associating domains, A/B compartmentalization, and enhancer-gene interactions all contriBute toWards gene regulation at a gloBal scale. Heart failure is a syndrome ii characterized, in part, By a dysregulated gene expression program. We hypothesized that chromatin structure Becomes deranged during heart failure, and We found this to Be the case at multiple levels of chromatin organization. -

Metalloproteinase Cleavage of Versican at The
METALLOPROTEINASE CLEAVAGE OF VERSICAN AT THE FIBROBLAST CELL SURFACE by Sean Bertram Maurice B.Kin., University of Calgary, 1999 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in The Faculty of Graduate Studies (Dental Science) THE UNIVERSITY OF BRITISH COLUMBIA October, 2009 © Sean Bertram Maurice, 2009 ABSTRACT Versican is a large aggregating proteoglycan expressed in the pericellular matrix of fibroblast cells. It is highly expressed during development and remodeling. The regulated synthesis and degradation of versican are associated with physiological remodeling. Versican is expressed in fibroproliferative lesions of human pulmonary fibrosis and atherosclerosis. Stromal expression of versican is associated with many forms of cancer and may be predictive of poor prognosis. Abnormal persistence of the versican-rich matrix may contribute to fibroproliferative and oncogenic processes. The process of versican degradation is not understood, but as versican is a pericellular molecule, physiological degradation likely involves cell surface-associated proteolysis. As such, the overarching hypothesis for this work is that regulated versican turnover involves the cell surface-associated metalloproteinases ADAMTS-2, MMP-2 and MT1-MMP, that are expressed in versican-rich remodeling lesions. ADAMTS-2 is a procollagen N-propeptidase involved in collagen fibrillogenesis. As procollagen is synthesized in a versican-rich matrix, it was hypothesized that ADAMTS- 2 might bind and process versican. MMP-2 and MT1-MMP in complex with TIMP-2, are activated at the cell surface during wound healing, pulmonary fibrosis and cancer. Versican was purified from human fetal lung fibroblast cultures for in vitro proteolysis experiments. The purified versican preparation was characterized by electrophoresis, chromatography, spectrophotometry and mass spectrometry. -

Evolutionary Convergence and Divergence in Archaeal
www.nature.com/scientificreports OPEN Evolutionary convergence and divergence in archaeal chromosomal proteins and Received: 30 November 2017 Accepted: 4 April 2018 Chromo-like domains from bacteria Published: xx xx xxxx and eukaryotes Gurmeet Kaur1, Lakshminarayan M. Iyer1, Srikrishna Subramanian2 & L. Aravind1 SH3-fold-β-barrel domains of the chromo-like superfamily recognize epigenetic marks in eukaryotic proteins. Their provenance has been placed either in archaea, based on apparent structural similarity to chromatin-compacting Sul7d and Cren7 proteins, or in bacteria based on the presence of sequence homologs. Using sequence and structural evidence we establish that the archaeal Cren7/Sul7 proteins emerged from a zinc ribbon (ZnR) ancestor. Further, we show that the ancestral eukaryotic chromo- like domains evolved from bacterial versions, likely acquired from early endosymbioses, which already possessed an aromatic cage for recognition of modifed amino-groups. These bacterial versions are part of a radiation of secreted SH3-fold domains, which spawned both chromo-like domains and classical SH3 domains in the context of peptide-recognition in the peptidoglycan or the extracellular matrix. This establishes that Cren7/Sul7 converged to a “SH3”-like state from a ZnR precursor via the loss of metal-chelation and acquisition of stronger hydrophobic interactions; it is unlikely to have participated in the evolution of the chromo-like domains. We show that archaea possess several Cren7/Sul7-related proteins with intact Zn-chelating ligands, which we predict to play previously unstudied roles in chromosome segregation during cell-division comparable to the PRC barrel and CdvA domain proteins. Tree-dimensional structures or folds of proteins are evolutionarily less prone to change than their sequences1–3.