Genetic Variation of Tunisian Myrtus Communis L. (Myrtaceae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Two New Taxa of Verticordia (Myrtaceae: Chamelaucieae) from South-Western Australia
A.S.Nuytsia George 20: 309–318 & M.D. (2010)Barrett,, Two new taxa of Verticordia 309 Two new taxa of Verticordia (Myrtaceae: Chamelaucieae) from south-western Australia Alex S. George1 and Matthew D. Barrett2,3 1 ‘Four Gables’, 18 Barclay Road, Kardinya, Western Australia 6163 Email: [email protected] 2 Botanic Gardens and Parks Authority, Kings Park and Botanic Garden, Fraser Ave, West Perth, Western Australia 6005 3 School of Plant Biology, University of Western Australia, Crawley, Western Australia 6009 Email: [email protected] Abstract George, A.S. and Barrett, M.D. Two new taxa of Verticordia (Myrtaceae: Chamelaucieae) from south- western Australia. Nuytsia 20: 309–318 (2010). Verticordia mitchelliana subsp. implexior A.S.George & M.D.Barrett and Verticordia setacea A.S.George are described and discussed. Verticordia setacea belongs with V. gracilis A.S.George in section Platandra, previously a monotypic section. Introduction The genus Verticordia DC. (Myrtaceae: tribe Chamelaucieae) is a charismatic group of shrubs found mainly in south-western Australia, with several species in adjacent arid regions and three in tropical Australia (George 1991; George & Pieroni 2002). Verticordia is currently defined solely on the possession of divided calyx lobes, but the limits between Verticordia and the related genera Homoranthus A.Cunn. ex Schauer, Chamelaucium Desf. and Darwinia Rudge are difficult to define conclusively, and other characteristics such as anther morphology suggest conflicting relationships (Bentham 1867; Craven & Jones 1991; George 1991). A recent analysis using a single chloroplast gene, with limited sampling of Verticordia taxa (Ma et al. 2002), suggests that Verticordia may be polyphyletic. -
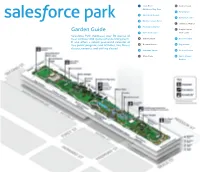
Salesforce Park Garden Guide
Start Here! D Central Lawn Children’s Play Area Garden Guide6 Palm Garden 1 Australian Garden Start Here! D Central Lawn Salesforce Park showcases7 California over Garden 50 species of Children’s Play Area 2 Mediterraneantrees and Basin over 230 species of understory plants. 6 Palm Garden -ã ¼ÜÊ ÊăØÜ ØÊèÜãE úØƀØÊèÃJapanese Maples ¼ÃØ Ê¢ 1 Australian Garden 3 Prehistoric¢ØÕ輫ÕØÊ£ØÂÜÃã«ó«ã«Üŧ¼«¹ĆãÃÜÜ Garden 7 California Garden ¼ÜÜÜŧÊÃØãÜŧÃØ¢ã«Ã£¼ÜÜÜũF Amphitheater Garden Guide 2 Mediterranean Basin 4 Wetland Garden Main Lawn E Japanese Maples Salesforce Park showcases over 50 species of 3 Prehistoric Garden trees and over 230 species of understory plants. A Oak Meadow 8 Desert Garden F Amphitheater It also offers a robust year-round calendar of 4 Wetland Garden Main Lawn free public programs and activities, like fitness B Bamboo Grove 9 Fog Garden Desert Garden classes, concerts, and crafting classes! A Oak Meadow 8 5 Redwood Forest 10 Chilean Garden B Bamboo Grove 9 Fog Garden C Main Plaza 11 South African 10 Chilean Garden Garden 5 Redwood Forest C Main Plaza 11 South African Garden 1 Children’s Australian Play Area Garden ABOUT THE GARDENS The botanist aboard the Endeavor, Sir Joseph Banks, is credited with introducing many plants from Australia to the western world, and many This 5.4 acre park has a layered soil system that plants today bear his name. balances seismic shifting, collects and filters storm- water, and irrigates the gardens. Additionally, the soil Native to eastern Australia, Grass Trees may grow build-up and dense planting help offset the urban only 3 feet in 100 years, and mature plants can be heat island effect by lowering the air temperature. -

The Biology of Casmara Subagronoma (Lepidoptera
insects Article The Biology of Casmara subagronoma (Lepidoptera: Oecophoridae), a Stem-Boring Moth of Rhodomyrtus tomentosa (Myrtaceae): Descriptions of the Previously Unknown Adult Female and Immature Stages, and Its Potential as a Biological Control Candidate Susan A. Wineriter-Wright 1, Melissa C. Smith 1,* , Mark A. Metz 2 , Jeffrey R. Makinson 3 , Bradley T. Brown 3, Matthew F. Purcell 3, Kane L. Barr 4 and Paul D. Pratt 5 1 USDA-ARS Invasive Plant Research Laboratory, Fort Lauderdale, FL 33314, USA; [email protected] 2 USDA-ARS Systematic Entomology Lab, Beltsville, MD 20013-7012, USA; [email protected] 3 USDA-ARS Australian Biological Control Laboratory, CSIRO Health and Biosecurity, Dutton Park QLD 4102, Australia; jeff[email protected] (J.R.M.); [email protected] (B.T.B.); [email protected] (M.F.P.) 4 USDA-ARS Center for Medical, Agricultural and Veterinary Entomology, Gainesville, FL 32608, USA; [email protected] 5 USDA-ARS, Western Regional Research Center, Invasive Species and Pollinator Health Research Unit, 800 Buchanan Street, Albany, CA 94710, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-954-475-6549 Received: 27 August 2020; Accepted: 16 September 2020; Published: 23 September 2020 Simple Summary: Rhodomyrtus tomentosa is a perennial woody shrub throughout Southeast Asia. Due to its prolific flower and fruit production, it was introduced into subtropical areas such as Florida and Hawai’i, where it is now naturalized and invasive. In an effort to find sustainable means to control R. tomentosa, a large-scale survey was mounted for biological control organisms. -

Lauraceae Along an Altitudinal Gradient in Southern Brazil
Floresta e Ambiente 2019; 26(4): e20170637 https://doi.org/10.1590/2179-8087.063717 ISSN 2179-8087 (online) Original Article Forest Management Lauraceae Along an Altitudinal Gradient in Southern Brazil Marcelo Leandro Brotto1 , Eduardo Damasceno Lozano2 , Felipe Eduardo Cordeiro Marinero2 , Alexandre Uhlmann3 , Christopher Thomas Blum2 , Carlos Vellozo Roderjan2 1Prefeitura Municipal de Curitiba, Curitiba, PR, Brasil 2Universidade Federal do Paraná (UFPR), Curitiba, PR, Brasil 3Embrapa Pesca e Aquicultura, Palmas, TO, Brasil ABSTRACT We performed a phytosociological study on an altitudinal gradient in Lauráceas State Park (Parque Estadual das Lauráceas/PR), aiming to describe the Montane Atlantic Rain Forest, to verify the importance of Lauraceae, and to evaluate the communities’ successional stage. We distributed survey units (2,000 m² quadrats) along an altitudinal gradient and surveyed all individuals with DBH ≥ 10 cm, which composed the arboreal component. In smaller quadrats (250 m²), we surveyed regeneration individuals. The community at 800 and 900 m a.s.l. shows typical characteristics of Montane forest in an advanced successional stage, and the abundance of Ocotea catharinensis is its main indicator. At 1,000 and 1,100 m a.s.l., the forest is characterized as Montane with short stature in an advanced successional stage, with the occurrence of typical upper montane species such as O. porosa and O. vaccinioides. Keywords: Atlantic forest, Lauráceas State Park, phytosociology. Creative Commons License. All the contents of this journal, except where otherwise noted, is licensed under a Creative Commons Attribution License. 2/12 Brotto ML, Lozano ED, Marinero FEC, Uhlmann A, Blum CT, Roderjan CV Floresta e Ambiente 2019; 26(4): e20170637 1. -

Myrtus Communis L.) Populations
Electronic Journal of Biotechnology ISSN: 0717-3458 http://www.ejbiotechnology.info DOI: 10.2225/vol16-issue6-fulltext-13 RESEARCH ARTICLE The genetic diversity of Sardinian myrtle (Myrtus communis L.) populations Sara Melito1 · Innocenza Chessa1 · Patrizia Erre1 · János Podani2 · Maurizio Mulas1 1 University of Sassari, Department of Science for Nature and Environmental Resources, Sassari, Italy 2 Eötvös University, Department of Plant Systematics, Ecology and Theoretical Biology, Budapest, Hungary Corresponding author: [email protected] Received May 2, 2013 / Accepted October 25, 2013 Published online: November 15, 2013 © 2013 by Pontificia Universidad Católica de Valparaíso, Chile Abstract Background: The myrtle (Myrtus communis) is a common shrub widespread in the Mediterranean Basin. Its fruit and leaves exhibit antioxidant, antibacterial and antifungal properties, and are used for their content of essential oils and for their medicinal properties, but most commonly as an ingredient in locally made liquor. The uncontrolled exploitation of natural stands has reduced both the species' geographical coverage and the size of individual populations. The selection of genotypes for controlled cultivation requires a characterization of the genetic diversity present both within and between populations. Results: Genotypic variation was evaluated using ISSR profiling and genetic diversity characterized using standard population genetics approaches. Two major clusters were identified: one capturing all the candidate cultivars selected from various Sardinian localities, and the other wild individuals collected from Asinara, Corsica and Surigheddu. A moderate level of gene flow between the Sardinian and Corsican populations was identified. Discriminant analysis of principal components revealed a level of separation among the wild populations, confirming the population structure identified by the clustering methods. -

Chemical Composition of Myrtus Communis L. (Myrtaceae) Fruits
Journal of Applied Life Sciences International 12(3): 1-8, 2017; Article no.JALSI.33746 ISSN: 2394-1103 Chemical Composition of Myrtus communis L. (Myrtaceae) Fruits Karzan Omer Qader1, Sahar A. A. Malik Al-Saadi2* and Thuraya A. Al-Saadi3 1Department of Biology, College of Science, Sulaimani University, Iraq. 2Department of Biology, College of Science, Basrah University, Iraq. 3Department of Science, College of Basic Education, Al-Mustansiriya University, Iraq. Authors’ contributions This work was carried out in collaboration between all authors. Authors KOQ and SAAMAS designed the study, performed sample collection and processing and wrote the first draft of the manuscript. Author TAAS wrote the protocol, managed the analyses of the study and managed the literature searches. All authors read and approved the final manuscript. Article Information DOI: 10.9734/JALSI/2017/33746 Editor(s): (1) Ali Mohamed Elshafei Ali, National Research Centre, Egypt. Reviewers: (1) Dildar Ahmed, Forman Christian College, Lahore, Pakistan. (2) Aidi Wannes Wissem, Center of Biotechnology of the Technopol Borj-Cedria, Tunisia. (3) Olabanji Iyabo Oluremi, Obafemi Awolowo University, Nigeria. Complete Peer review History: http://www.sciencedomain.org/review-history/19748 Received 27th April 2017 Accepted 17th June 2017 Original Research Article th Published 28 June 2017 ABSTRACT The chemical composition of Myrtus communis L. extracts were prepared and analyzed by GC-MS. Sixteen phytochemical constituents of chemical compounds were identified in fruits of Myrtus communis. The relative percentage of linoleic acid methyl ester was high (27.19%), followed by oleic acid methyl ester (21.18%) and then octane 3,5- dimethyl (16.47%), dodecane (11.39%), palmitic acid methyl ester (6.80%) and tetradecane (6.69%) as well as, some components present in lower percentage such as stearic acid methyl ester (3.32%). -

American Magazine
The American HORTICULTURAL Magazine spring 1970 / volume 49 / number 2 Journal of the American Horticultural Society, Inc. 2401 CALVERT STREET, N.W. / WASHINGTON, D. c. 20008 For United Horticulture . The particular objec.ts and business of the American Horticultural Society are to promote and encourage national in·terest in scientific research and education in horticulture in all of its branches. 1969-1970 EXEOUTIVE COMMI'TTEE* President Secretary MR. FRED C. GALLE (1970) MRS. FRANCIS PATTESON-KNIGHT (1970) Director of Horticulture Hidden Acres Callaway Gardens 8607 Tebbs Lane Pine Mountain, Georgia 31822 McLean, Virgini.a 22101 Treasurer and Immediate Past President First Vice President MR. JOHN H. WALKER (1970) DR. DAVID G. LEACH (1970) Execu.tive Director 1674 Trinity Road The Society of American Florists North Madison, Ohio 44057 901 North Washington Street Alexandria, Virginia 223-14 Second Vice President DR. NEIL W. STUART (1970) Member of the Board Plant Physiologist DR. HAROLD B. TUKEY, SR. (1970) Crops Research Division Professor Emeritus Agricultural Research Service Michigan State University U. S. Department of Agriculture The Maples Beltsville, Maryland 20705 Woodland, Michigan 48897 Assistant Treasurer Assistant Secretary MR. GLENN B. EASTBURN MRS. ELIZABETH G. EASTBURN Finance Officer Executive Director Washington, D. C. Washington, D. C. • Mem.bers of the 1969·70 Board of Directors per bylaw provision. THE AMERICAN HORTICULTURAL MAGAZINE is the official publication of The American Horticultural Society and is issued during the Winter, Spring, Summer, and Fall quarters. The magazine is included as a benefit of membership in The American Horticultural Society, individual membership dues being $15.00 a year. -

Darwinia Hortiorum (Myrtaceae: Chamelaucieae), a New Species from the Darling Range, Western Australia
K.R.Nuytsia Thiele, 20: 277–281 Darwinia (2010) hortiorum (Myrtaceae: Chamelaucieae), a new species 277 Darwinia hortiorum (Myrtaceae: Chamelaucieae), a new species from the Darling Range, Western Australia Kevin R. Thiele Western Australian Herbarium, Department of Environment and Conservation, Locked Bag 104, Bentley Delivery Centre, Western Australia 6983 Email: [email protected] Abstract Thiele, K.R. Darwinia hortiorum (Myrtaceae: Chamelaucieae), a new species from the Darling Range, Western Australia. Nuytsia 20: 277–281 (2010). The distinctive, new, rare species Darwinia hortiorum is described, illustrated and discussed. Uniquely in the genus it has strongly curved- zygomorphic flowers with the sigmoid styles arranged so that they group towards the centre of the head-like inflorescences. Introduction Darwinia Rudge comprises c. 90 species, mostly from the south-west of Western Australia with c. 15 species in New South Wales, Victoria and South Australia. Phylogenetic analyses (M. Barrett, unpublished) have shown that the genus is polyphyletic, with distinct eastern and western Australian clades. Along with the related genera Actinodium Schauer, Chamelaucium Desf., Homoranthus A.Cunn. ex Schauer and Pileanthus Labill., the Darwinia clades are nested in a paraphyletic Verticordia DC. Many undescribed species of Darwinia are known in Western Australia, and these are being progressively described (Rye 1983; Marchant & Keighery 1980; Marchant 1984; Keighery & Marchant 2002; Keighery 2009). A significant number of taxa in the genus are narrowly endemic or rare and are of high conservation significance. Although taxonomic reassignment of the Western Australian species of Darwinia may be required in the future, resolving the status of these undescribed species and describing them under their current genus helps provide information for conservation assessments and survey. -

Structures and Bioactive Properties of Myrtucommulones and Related Acylphloroglucinols from Myrtaceae
molecules Review Structures and Bioactive Properties of Myrtucommulones and Related Acylphloroglucinols from Myrtaceae Rosario Nicoletti 1,2 , Maria Michela Salvatore 3 , Pasquale Ferranti 2 and Anna Andolfi 3,* 1 Council for Agricultural Research and Economics, Research Centre for Olive, Citrus and Tree Fruit, 81100 Caserta, Italy; [email protected] 2 Department of Agriculture, University of Naples ‘Federico II’, 80055 Portici, Italy; [email protected] 3 Department of Chemical Sciences, University of Naples ‘Federico II’, 80126 Naples, Italy; [email protected] * Correspondence: andolfi@unina.it; Tel.: +39-081-2539179 Academic Editors: Francesco Vinale and Maria Luisa Balestrieri Received: 2 December 2018; Accepted: 17 December 2018; Published: 19 December 2018 Abstract: Myrtaceae are a group of plants that include a number of renowned species used in ethnomedicine in many areas worldwide. Their valuable therapeutic properties have stimulated a fruitful research activity addressed to the identification of the bioactive components of their extracts yielding a great diversity of terpenes; polyphenols; and other exclusive products. Among the latter, starting with the discovery of myrtucommulone A from myrtle (Myrtus communis), a series of structurally-related acylphloroglucinol compounds have been characterized from several species that represent the basic active principles to be considered in view of possible drug development. Aspects concerning chemical and biological properties of these products are reviewed in the present paper. Keywords: myrtucommulone; acylphloroglucinols; Myrtaceae; plant extracts; biological activities 1. Introduction Myrtle (Myrtus communis) is a typical shrub of maquis and coastal bushes native of the Mediterranean area and Western Asia. It is well-known in traditional medicine, and for centuries its leaves and berries have found ethnomedical application in the treatment of several disorders of the digestive apparatus, as well as pulmonary and skin diseases [1,2]. -

Morphology and Vascular Anatomy of the Flower of Angophora Intermedia
© Landesmuseum für Kärnten; download www.landesmuseum.ktn.gv.at/wulfenia; www.biologiezentrum.at Wulfenia 13 (2006): 11–19 Mitteilungen des Kärntner Botanikzentrums Klagenfurt Morphology and vascular anatomy of the fl ower of Angophora intermedia DC. (Myrtaceae) with special emphasis on the innervation of the fl oral axis Sergey A. Volgin & Anastasiya Stepanova Summary: A peculiar receptacle structure in Angophora intermedia DC. (Myrtaceae) has been determined by a vascular-anatomical method. The vascular system of the fl ower of A. intermedia consists of numerous ascending bundles and girdling bundles in the hypanthium and the inferior ovary wall. In the central column of the trilocular ovary we found a dense conical plexus of vascular bundles supplying the placentae (infralocular plexus). It is connected with ascending bundles of the receptacle in the ovary base. In its central part it contains “hanged” bundles and blind bundles, so it seems to be a residual stele of a rudimentary fl oral apex. Thus, the receptacle ofA. intermedia is toroidal at the level of fl oral organs and conical above the carpel node. Keywords: Angophora intermedia, Myrtaceae, fl ower morphology, vascular system, fl oral axis, innervation, anatomy The fl oral development in different species of Myrtaceae has been studied precisely to elucidate the homology of the inferior ovary, hypanthium, operculate perianth and stamens of the polymerous androecium (PAYER 1857; MAYR 1969; BUNNIGER 1972; DRINNAN & LADIGES 1988; RONSE DECRAENE & SMETS 1991; ORLOVICH et al. 1996). Developmental and histogenetical studies have shown, that the receptacle in the fl ower of Myrtaceae is cup-like and take part to certain extent in the formation of the inferior ovary wall and the hypanthium (PAYER 1857; BUNNIGER 1972; RONSE DECRAENE & SMETS 1991). -

Vascular Plant Families of the United States Grouped by Diagnostic Features
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 12-6-2019 Vascular Plant Families of the United States Grouped by Diagnostic Features James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Vascular Plant Families of the United States Grouped by Diagnostic Features" (2019). Botanical Studies. 96. https://digitalcommons.humboldt.edu/botany_jps/96 This Flora of the United States and North America is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. FLOWERING PLANT FAMILIES OF THE UNITED STATES GROUPED BY DIAGNOSTIC FEATURES James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State University Second edition — 6 December 2019 The focus is on families of plants found in the conterminous United States, including ornamentals. The listing of a family is not meant to imply that every species has that feature. I am using a fewfamily names, such as Liliaceae, Plantaginaceae, and Scrophulariaceae, in the traditional sense, because their limits remain unsettled. Parasitic on branches Dioscoreaceae -

Genera in Myrtaceae Family
Genera in Myrtaceae Family Genera in Myrtaceae Ref: http://data.kew.org/vpfg1992/vascplnt.html R. K. Brummitt 1992. Vascular Plant Families and Genera, Royal Botanic Gardens, Kew REF: Australian – APC http://www.anbg.gov.au/chah/apc/index.html & APNI http://www.anbg.gov.au/cgi-bin/apni Some of these genera are not native but naturalised Tasmanian taxa can be found at the Census: http://tmag.tas.gov.au/index.aspx?base=1273 Future reference: http://tmag.tas.gov.au/floratasmania [Myrtaceae is being edited at mo] Acca O.Berg Euryomyrtus Schaur Osbornia F.Muell. Accara Landrum Feijoa O.Berg Paragonis J.R.Wheeler & N.G.Marchant Acmena DC. [= Syzigium] Gomidesia O.Berg Paramyrciaria Kausel Acmenosperma Kausel [= Syzigium] Gossia N.Snow & Guymer Pericalymma (Endl.) Endl. Actinodium Schauer Heteropyxis Harv. Petraeomyrtus Craven Agonis (DC.) Sweet Hexachlamys O.Berg Phymatocarpus F.Muell. Allosyncarpia S.T.Blake Homalocalyx F.Muell. Pileanthus Labill. Amomyrtella Kausel Homalospermum Schauer Pilidiostigma Burret Amomyrtus (Burret) D.Legrand & Kausel [=Leptospermum] Piliocalyx Brongn. & Gris Angasomyrtus Trudgen & Keighery Homoranthus A.Cunn. ex Schauer Pimenta Lindl. Angophora Cav. Hottea Urb. Pleurocalyptus Brongn. & Gris Archirhodomyrtus (Nied.) Burret Hypocalymma (Endl.) Endl. Plinia L. Arillastrum Pancher ex Baill. Kania Schltr. Pseudanamomis Kausel Astartea DC. Kardomia Peter G. Wilson Psidium L. [naturalised] Asteromyrtus Schauer Kjellbergiodendron Burret Psiloxylon Thouars ex Tul. Austromyrtus (Nied.) Burret Kunzea Rchb. Purpureostemon Gugerli Babingtonia Lindl. Lamarchea Gaudich. Regelia Schauer Backhousia Hook. & Harv. Legrandia Kausel Rhodamnia Jack Baeckea L. Lenwebia N.Snow & ZGuymer Rhodomyrtus (DC.) Rchb. Balaustion Hook. Leptospermum J.R.Forst. & G.Forst. Rinzia Schauer Barongia Peter G.Wilson & B.Hyland Lindsayomyrtus B.Hyland & Steenis Ristantia Peter G.Wilson & J.T.Waterh.