FARE2015 WINNERS Sorted by Institute/Center
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Compensation Among HMGN Variants Modulates the Dnase I Hypersensitive Sites at Enhancers
Downloaded from genome.cshlp.org on October 9, 2021 - Published by Cold Spring Harbor Laboratory Press Research Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers Tao Deng,1,12 Z. Iris Zhu,2,12 Shaofei Zhang,1 Yuri Postnikov,1 Di Huang,2 Marion Horsch,3 Takashi Furusawa,1 Johannes Beckers,3,4,5 Jan Rozman,3,5 Martin Klingenspor,6,7 Oana Amarie,3,8 Jochen Graw,3,8 Birgit Rathkolb,3,5,9 Eckhard Wolf,9 Thure Adler,3 Dirk H. Busch,10 Valérie Gailus-Durner,3 Helmut Fuchs,3 Martin Hrabeˇ de Angelis,3,4,5 Arjan van der Velde,2,13 Lino Tessarollo,11 Ivan Ovcherenko,2 David Landsman,2 and Michael Bustin1 1Protein Section, Laboratory of Metabolism, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892, USA; 2Computational Biology Branch, National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland 20892, USA; 3German Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, German Research Center for Environmental Health, 85764 Neuherberg, Germany; 4Experimental Genetics, Center of Life and Food Sciences Weihenstephan, Technische Universität München, 85354 Freising-Weihenstephan, Germany; 5German Center for Diabetes Research (DZD), 85764 Neuherberg, Germany; 6Molecular Nutritional Medicine, Technische Universität München, 85350 Freising, Germany; 7Center for Nutrition and Food Sciences, Technische Universität München, 85350 Freising, Germany; 8Institute of Developmental Genetics (IDG), 85764 -

From Inverse Agonism to 'Paradoxical Pharmacology' Richard A
International Congress Series 1249 (2003) 27-37 From inverse agonism to 'Paradoxical Pharmacology' Richard A. Bond*, Kenda L.J. Evans, Zsirzsanna Callaerts-Vegh Department of Pharmacological and Pharmaceutical Sciences, University of Houston, 521 Science and Research Bldg 2, 4800 Caltioun, Houston, TX 77204-5037, USA Received 16 April 2003; accepted 16 April 2003 Abstract The constitutive or spontaneous activity of G protein-coupled receptors (GPCRs) and compounds acting as inverse agonists is a recent but well-established phenomenon. Dozens of receptor subtypes for numerous neurotransmitters and hormones have been shown to posses this property. However, do to the apparently low percentage of receptors in the spontaneously active state, the physiologic relevance of these findings remains questionable. The possibility that the reciprocal nature of the effects of agonists and inverse agonists may extend to cellular signaling is discussed, and that this may account for the beneficial effects of certain p-adrenoceptor inverse agonists in the treatment of heart failure. © 2003 Elsevier Science B.V. All rights reserved. Keywords. Inverse agonism; GPCR; Paradoxical pharmacology 1. Brief history of inverse agonism at G protein-coupled receptors For approximately three-quarters of a century, ligands that interacted with G protein- coupled receptors (GPCRs) were classified either as agonists or antagonists. Receptors were thought to exist in a single quiescent state that could only induce cellular signaling upon agonist binding to the receptor to produce an activated state of the receptor. In this model, antagonists had no cellular signaling ability on their own, but did bind to the receptor and prevented agonists from being able to bind and activate the receptor. -

Prox1regulates the Subtype-Specific Development of Caudal Ganglionic
The Journal of Neuroscience, September 16, 2015 • 35(37):12869–12889 • 12869 Development/Plasticity/Repair Prox1 Regulates the Subtype-Specific Development of Caudal Ganglionic Eminence-Derived GABAergic Cortical Interneurons X Goichi Miyoshi,1 Allison Young,1 Timothy Petros,1 Theofanis Karayannis,1 Melissa McKenzie Chang,1 Alfonso Lavado,2 Tomohiko Iwano,3 Miho Nakajima,4 Hiroki Taniguchi,5 Z. Josh Huang,5 XNathaniel Heintz,4 Guillermo Oliver,2 Fumio Matsuzaki,3 Robert P. Machold,1 and Gord Fishell1 1Department of Neuroscience and Physiology, NYU Neuroscience Institute, Smilow Research Center, New York University School of Medicine, New York, New York 10016, 2Department of Genetics & Tumor Cell Biology, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105, 3Laboratory for Cell Asymmetry, RIKEN Center for Developmental Biology, Kobe 650-0047, Japan, 4Laboratory of Molecular Biology, Howard Hughes Medical Institute, GENSAT Project, The Rockefeller University, New York, New York 10065, and 5Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724 Neurogliaform (RELNϩ) and bipolar (VIPϩ) GABAergic interneurons of the mammalian cerebral cortex provide critical inhibition locally within the superficial layers. While these subtypes are known to originate from the embryonic caudal ganglionic eminence (CGE), the specific genetic programs that direct their positioning, maturation, and integration into the cortical network have not been eluci- dated. Here, we report that in mice expression of the transcription factor Prox1 is selectively maintained in postmitotic CGE-derived cortical interneuron precursors and that loss of Prox1 impairs the integration of these cells into superficial layers. Moreover, Prox1 differentially regulates the postnatal maturation of each specific subtype originating from the CGE (RELN, Calb2/VIP, and VIP). -

Clinical Utility of Recently Identified Diagnostic, Prognostic, And
Modern Pathology (2017) 30, 1338–1366 1338 © 2017 USCAP, Inc All rights reserved 0893-3952/17 $32.00 Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms Arantza Onaindia1, L Jeffrey Medeiros2 and Keyur P Patel2 1Instituto de Investigacion Marques de Valdecilla (IDIVAL)/Hospital Universitario Marques de Valdecilla, Santander, Spain and 2Department of Hematopathology, MD Anderson Cancer Center, Houston, TX, USA Genomic profiling studies have provided new insights into the pathogenesis of mature B-cell neoplasms and have identified markers with prognostic impact. Recurrent mutations in tumor-suppressor genes (TP53, BIRC3, ATM), and common signaling pathways, such as the B-cell receptor (CD79A, CD79B, CARD11, TCF3, ID3), Toll- like receptor (MYD88), NOTCH (NOTCH1/2), nuclear factor-κB, and mitogen activated kinase signaling, have been identified in B-cell neoplasms. Chronic lymphocytic leukemia/small lymphocytic lymphoma, diffuse large B-cell lymphoma, follicular lymphoma, mantle cell lymphoma, Burkitt lymphoma, Waldenström macroglobulinemia, hairy cell leukemia, and marginal zone lymphomas of splenic, nodal, and extranodal types represent examples of B-cell neoplasms in which novel molecular biomarkers have been discovered in recent years. In addition, ongoing retrospective correlative and prospective outcome studies have resulted in an enhanced understanding of the clinical utility of novel biomarkers. This progress is reflected in the 2016 update of the World Health Organization classification of lymphoid neoplasms, which lists as many as 41 mature B-cell neoplasms (including provisional categories). Consequently, molecular genetic studies are increasingly being applied for the clinical workup of many of these neoplasms. In this review, we focus on the diagnostic, prognostic, and/or therapeutic utility of molecular biomarkers in mature B-cell neoplasms. -

The Structure and Antioxidant Properties
materials Review Recent Developments in Effective Antioxidants: The Structure and Antioxidant Properties Monika Parcheta 1 , Renata Swisłocka´ 1,* , Sylwia Orzechowska 2,3 , Monika Akimowicz 4 , Renata Choi ´nska 4 and Włodzimierz Lewandowski 1 1 Department of Chemistry, Biology and Biotechnology, Bialystok University of Technology, Wiejska 45E, 15-351 Bialystok, Poland; [email protected] (M.P.); [email protected] (W.L.) 2 Solaris National Synchrotron Radiation Centre, Jagiellonian University, Czerwone Maki 98, 30-392 Krakow, Poland; [email protected] 3 M. Smoluchowski Institute of Physics, Jagiellonian University, Łojasiewicza 11, 30-348 Kraków, Poland 4 Prof. Waclaw Dabrowski Institute of Agriculture and Food Biotechnology–State Research Institute, Rakowiecka 36, 02-532 Warsaw, Poland; [email protected] (M.A.); [email protected] (R.C.) * Correspondence: [email protected] Abstract: Since the last few years, the growing interest in the use of natural and synthetic antioxidants as functional food ingredients and dietary supplements, is observed. The imbalance between the number of antioxidants and free radicals is the cause of oxidative damages of proteins, lipids, and DNA. The aim of the study was the review of recent developments in antioxidants. One of the crucial issues in food technology, medicine, and biotechnology is the excess free radicals reduction to obtain healthy food. The major problem is receiving more effective antioxidants. The study aimed to analyze the properties of efficient antioxidants and a better understanding of the molecular ´ Citation: Parcheta, M.; Swisłocka, R.; mechanism of antioxidant processes. Our researches and sparing literature data prove that the Orzechowska, S.; Akimowicz, M.; ligand antioxidant properties complexed by selected metals may significantly affect the free radical Choi´nska,R.; Lewandowski, W. -

Genome-Wide DNA Methylation Analysis of KRAS Mutant Cell Lines Ben Yi Tew1,5, Joel K
www.nature.com/scientificreports OPEN Genome-wide DNA methylation analysis of KRAS mutant cell lines Ben Yi Tew1,5, Joel K. Durand2,5, Kirsten L. Bryant2, Tikvah K. Hayes2, Sen Peng3, Nhan L. Tran4, Gerald C. Gooden1, David N. Buckley1, Channing J. Der2, Albert S. Baldwin2 ✉ & Bodour Salhia1 ✉ Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 diferentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in diferentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream efector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer. Activating KRAS mutations can be found in nearly 25 percent of all cancers1. -

HMGB1 in Health and Disease R
Donald and Barbara Zucker School of Medicine Journal Articles Academic Works 2014 HMGB1 in health and disease R. Kang R. C. Chen Q. H. Zhang W. Hou S. Wu See next page for additional authors Follow this and additional works at: https://academicworks.medicine.hofstra.edu/articles Part of the Emergency Medicine Commons Recommended Citation Kang R, Chen R, Zhang Q, Hou W, Wu S, Fan X, Yan Z, Sun X, Wang H, Tang D, . HMGB1 in health and disease. 2014 Jan 01; 40():Article 533 [ p.]. Available from: https://academicworks.medicine.hofstra.edu/articles/533. Free full text article. This Article is brought to you for free and open access by Donald and Barbara Zucker School of Medicine Academic Works. It has been accepted for inclusion in Journal Articles by an authorized administrator of Donald and Barbara Zucker School of Medicine Academic Works. Authors R. Kang, R. C. Chen, Q. H. Zhang, W. Hou, S. Wu, X. G. Fan, Z. W. Yan, X. F. Sun, H. C. Wang, D. L. Tang, and +8 additional authors This article is available at Donald and Barbara Zucker School of Medicine Academic Works: https://academicworks.medicine.hofstra.edu/articles/533 NIH Public Access Author Manuscript Mol Aspects Med. Author manuscript; available in PMC 2015 December 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Mol Aspects Med. 2014 December ; 0: 1–116. doi:10.1016/j.mam.2014.05.001. HMGB1 in Health and Disease Rui Kang1,*, Ruochan Chen1, Qiuhong Zhang1, Wen Hou1, Sha Wu1, Lizhi Cao2, Jin Huang3, Yan Yu2, Xue-gong Fan4, Zhengwen Yan1,5, Xiaofang Sun6, Haichao Wang7, Qingde Wang1, Allan Tsung1, Timothy R. -

Making Sense of Pharmacology: Inverse Agonism and Functional Selectivity Kelly A
International Journal of Neuropsychopharmacology (2018) 21(10): 962–977 doi:10.1093/ijnp/pyy071 Advance Access Publication: August 6, 2018 Review review Making Sense of Pharmacology: Inverse Agonism and Functional Selectivity Kelly A. Berg and William P. Clarke Department of Pharmacology, University of Texas Health, San Antonio, Texas. Correspondence: William P. Clarke, PhD, Department of Pharmacology, Mail Stop 7764, UT Health at San Antonio, 7703 Floyd Curl Drive, San Antonio, TX 78229 ([email protected]). Abstract Constitutive receptor activity/inverse agonism and functional selectivity/biased agonism are 2 concepts in contemporary pharmacology that have major implications for the use of drugs in medicine and research as well as for the processes of new drug development. Traditional receptor theory postulated that receptors in a population are quiescent unless activated by a ligand. Within this framework ligands could act as agonists with various degrees of intrinsic efficacy, or as antagonists with zero intrinsic efficacy. We now know that receptors can be active without an activating ligand and thus display “constitutive” activity. As a result, a new class of ligand was discovered that can reduce the constitutive activity of a receptor. These ligands produce the opposite effect of an agonist and are called inverse agonists. The second topic discussed is functional selectivity, also commonly referred to as biased agonism. Traditional receptor theory also posited that intrinsic efficacy is a single drug property independent of the system in which the drug acts. However, we now know that a drug, acting at a single receptor subtype, can have multiple intrinsic efficacies that differ depending on which of the multiple responses coupled to a receptor is measured. -

Cannabinoid-1 Receptor Inverse Agonists: Current Understanding of Mechanism of Action and Unanswered Questions
International Journal of Obesity (2009) 33, 947–955 & 2009 Macmillan Publishers Limited All rights reserved 0307-0565/09 $32.00 www.nature.com/ijo REVIEW Cannabinoid-1 receptor inverse agonists: current understanding of mechanism of action and unanswered questions TM Fong1 and SB Heymsfield2 1Merck Research Laboratories, Department of Metabolic Disorders, Rahway, NJ, USA and 2Merck Research Laboratories, Global Center of Scientific Affairs, Rahway, NJ, USA Rimonabant and taranabant are two extensively studied cannabinoid-1 receptor (CB1R) inverse agonists. Their effects on in vivo peripheral tissue metabolism are generally well replicated. The central nervous system site of action of taranabant or rimonabant is firmly established based on brain receptor occupancy studies. At the whole-body level, the mechanism of action of CB1R inverse agonists includes a reduction in food intake and an increase in energy expenditure. At the tissue level, fat mass reduction, liver lipid reduction and improved insulin sensitivity have been shown. These effects on tissue metabolism are readily explained by CB1R inverse agonist acting on brain CB1R and indirectly influencing the tissue metabolism through the autonomic nervous system. It has also been hypothesized that rimonabant acts directly on adipocytes, hepatocytes, pancreatic islets or skeletal muscle in addition to acting on brain CB1R, although strong support for the contribution of peripherally located CB1R to in vivo efficacy is still lacking. This review will carefully examine the published literature -
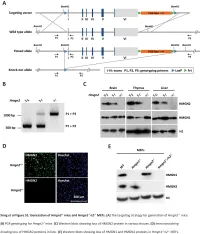
Supplemental Data.Pdf
Table S1. Summary of sequencing results A. DNase‐seq data % Align % Mismatch % >=Q30 bases Sample ID # Reads % unique reads (PF) Rate (PF) (PF) WT_1 84,301,522 86.76 0.52 93.34 96.30 WT_2 98,744,222 84.97 0.51 93.75 89.94 Hmgn1‐/‐_1 79,620,656 83.94 0.86 90.58 88.65 Hmgn1‐/‐_2 62,673,782 84.13 0.87 91.30 89.18 Hmgn2‐/‐_1 87,734,440 83.49 0.71 91.81 90.00 Hmgn2‐/‐_2 82,498,808 83.25 0.69 92.73 90.66 Hmgn1‐/‐n2‐/‐_1 71,739,638 68.51 2.31 81.11 89.22 Hmgn1‐/‐n2‐/‐_2 74,113,682 68.19 2.37 81.16 86.57 B. ChIP‐seq data Histone % Align % Mismatch % >=Q30 % unique Genotypes # Reads marks (PF) Rate (PF) bases (PF) reads H3K4me1 100670054 92.99 0.28 91.21 87.29 H3K4me3 67064272 91.97 0.35 89.11 27.15 WT H3K27ac 90,340,242 93.57 0.28 95.02 89.80 input 111,292,572 78.24 0.55 96.07 86.99 H3K4me1 84598176 92.34 0.33 91.2 81.69 H3K4me3 90032064 92.19 0.44 88.76 15.81 Hmgn1‐/‐n2‐/‐ H3K27ac 86,260,526 93.40 0.29 94.94 87.49 input 78,142,334 78.47 0.56 95.82 81.11 C. MNase‐seq data % Mismatch % >=Q30 bases % unique Sample ID # Reads % Align (PF) Rate (PF) (PF) reads WT_1_Extensive 45,232,694 55.23 1.49 90.22 81.73 WT_1_Limited 105,460,950 58.03 1.39 90.81 79.62 WT_2_Extensive 40,785,338 67.34 1.06 89.76 89.60 WT_2_Limited 105,738,078 68.34 1.05 90.29 85.96 Hmgn1‐/‐n2‐/‐_1_Extensive 117,927,050 55.74 1.49 89.50 78.01 Hmgn1‐/‐n2‐/‐_1_Limited 61,846,742 63.76 1.22 90.57 84.55 Hmgn1‐/‐n2‐/‐_2_Extensive 137,673,830 60.04 1.30 89.28 78.99 Hmgn1‐/‐n2‐/‐_2_Limited 45,696,614 62.70 1.21 90.71 85.52 D. -

At the X-Roads of Sex and Genetics in Pulmonary Arterial Hypertension
G C A T T A C G G C A T genes Review At the X-Roads of Sex and Genetics in Pulmonary Arterial Hypertension Meghan M. Cirulis 1,2,* , Mark W. Dodson 1,2, Lynn M. Brown 1,2, Samuel M. Brown 1,2, Tim Lahm 3,4,5 and Greg Elliott 1,2 1 Division of Pulmonary, Critical Care and Occupational Medicine, University of Utah, Salt Lake City, UT 84132, USA; [email protected] (M.W.D.); [email protected] (L.M.B.); [email protected] (S.M.B.); [email protected] (G.E.) 2 Division of Pulmonary and Critical Care Medicine, Intermountain Medical Center, Salt Lake City, UT 84107, USA 3 Division of Pulmonary, Critical Care, Sleep and Occupational Medicine, Department of Medicine, Indiana University School of Medicine, Indianapolis, IN 46202, USA; [email protected] 4 Richard L. Roudebush Veterans Affairs Medical Center, Indianapolis, IN 46202, USA 5 Department of Anatomy, Cell Biology & Physiology, Indiana University School of Medicine, Indianapolis, IN 46202, USA * Correspondence: [email protected]; Tel.: +1-801-581-7806 Received: 29 September 2020; Accepted: 17 November 2020; Published: 20 November 2020 Abstract: Group 1 pulmonary hypertension (pulmonary arterial hypertension; PAH) is a rare disease characterized by remodeling of the small pulmonary arteries leading to progressive elevation of pulmonary vascular resistance, ultimately leading to right ventricular failure and death. Deleterious mutations in the serine-threonine receptor bone morphogenetic protein receptor 2 (BMPR2; a central mediator of bone morphogenetic protein (BMP) signaling) and female sex are known risk factors for the development of PAH in humans. -

Myt1l Safeguards Neuronal Identity by Actively Repressing Many Non-Neuronal Fates Moritz Mall1, Michael S
LETTER doi:10.1038/nature21722 Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates Moritz Mall1, Michael S. Kareta1†, Soham Chanda1,2, Henrik Ahlenius3, Nicholas Perotti1, Bo Zhou1,2, Sarah D. Grieder1, Xuecai Ge4†, Sienna Drake3, Cheen Euong Ang1, Brandon M. Walker1, Thomas Vierbuchen1†, Daniel R. Fuentes1, Philip Brennecke5†, Kazuhiro R. Nitta6†, Arttu Jolma6, Lars M. Steinmetz5,7, Jussi Taipale6,8, Thomas C. Südhof2 & Marius Wernig1 Normal differentiation and induced reprogramming require human Myt1l (Extended Data Fig. 1). Chromatin immunoprecipita- the activation of target cell programs and silencing of donor cell tion followed by DNA sequencing (ChIP–seq) of endogenous Myt1l programs1,2. In reprogramming, the same factors are often used to in fetal neurons (embryonic day (E) 13.5) and ectopic Myt1l in mouse reprogram many different donor cell types3. As most developmental embryonic fibroblasts (MEFs) two days after induction identified 3,325 repressors, such as RE1-silencing transcription factor (REST) and high-confidence Myt1l peaks that overlapped remarkably well between Groucho (also known as TLE), are considered lineage-specific neurons and MEFs (Fig. 1a, Extended Data Fig. 2, Supplementary repressors4,5, it remains unclear how identical combinations of Table 1). Thus, similar to the pioneer factor Ascl1, Myt1l can access transcription factors can silence so many different donor programs. the majority of its cognate DNA binding sites even in a distantly related Distinct lineage repressors would have to be induced in different cell type. However, unlike Ascl1 targets8, the chromatin at Myt1l donor cell types. Here, by studying the reprogramming of mouse fibroblasts to neurons, we found that the pan neuron-specific a Myt1l Myt1l b Myt1l Ascl1 Random Myt1l 6 Ascl1 + Brn2 endogenous transcription factor Myt1-like (Myt1l) exerts its pro-neuronal Closed 0.030 function by direct repression of many different somatic lineage k programs except the neuronal program.