List of Plant Species Found in 2011 at BADL
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Threatened and Endangered Species Coorespondence
APPENDIXF THREATENED AND ENDANGERED SPECIES CORRESPONDENCE INTRODUCTION Forest Service policy regarding Biological Evaluaitons is summarized in Forest Service Manual (FSM) 2672.4. The intent of the Bilogical Evaluation process is to assess the potential impacts of propsed management activities, and ensure that such activities will not jeopardize the continued existence of: 1. Species listed, or proposed to be listed, as Engangered or Threatened by the U. S. Fish and Wildlife Service and 2. Species designated as sensitive by the Regional Forester. AFFECTED ENVIRONMENT Project Area The Sioux Ranger District proposes to update allotment management plans for 11 domestic livestock allotments on National Forest System lands in the North and South Cave Hills and East Short Pine land units. The decision associated with this proposal and analysis wi" determine where livestock can graze, when grazing wi" occur and what specific guidelines will be established to regulate the intensity (timing and duration) of grazing. The analysis area includes about 17,700 National Forest acres. The climate is continental and semi-arid, with large seasonal and daily temperature variations being common. Most of the rainfall during the summer is from thunderstorms; flash flooding can occur form the more severe thunderstorms. Streamflow is erratic with most streams being intermittant in nature. Ecological units include hardwood draws, Ponderosa bench and slope, upland, rolling, and table top grassland, rockland, and rimrocks. Detailed information regarding plant community composition and location is found elsewhere in the environmental analysis for this proposed action. Field surveys have been conducted within or adjacent to the project area by Linda Spencer, Kim Reid, Jeff DiBenedetto, and Tim McGarvey during the 2001 field season (s). -

Eriogonum Visheri A
Eriogonum visheri A. Nelson (Visher’s buckwheat): A Technical Conservation Assessment Prepared for the USDA Forest Service, Rocky Mountain Region, Species Conservation Project December 18, 2006 Juanita A. R. Ladyman, Ph.D. JnJ Associates LLC 6760 S. Kit Carson Cir E. Centennial, CO 80122 Peer Review Administered by Center for Plant Conservation Ladyman, J.A.R. (2006, December 18). Eriogonum visheri A. Nelson (Visher’s buckwheat): a technical conservation assessment. [Online]. USDA Forest Service, Rocky Mountain Region. Available: http://www.fs.fed.us/r2/ projects/scp/assessments/eriogonumvisheri.pdf [date of access]. ACKNOWLEDGMENTS The time spent and help given by all the people and institutions listed in the reference section are gratefully acknowledged. I would also like to thank the North Dakota Parks and Recreation Department, in particular Christine Dirk, and the South Dakota Natural Heritage Program, in particular David Ode, for their generosity in making their records, reports, and photographs available. I thank the Montana Natural Heritage Program, particularly Martin Miller, Mark Gabel of the Black Hills University Herbarium, Robert Tatina of the Dakota Wesleyan University, Christine Niezgoda of the Field Museum of Natural History, Carrie Kiel Academy of Natural Sciences, Dave Dyer of the University of Montana Herbarium, Caleb Morse of the R.L. McGregor Herbarium, Robert Kaul of the C. E. Bessey Herbarium, John La Duke of the University of North Dakota Herbarium, Joe Washington of the Dakota National Grasslands, and Doug Sargent of the Buffalo Gap National Grasslands - Region 2, for the information they provided. I also appreciate the access to files and assistance given to me by Andrew Kratz, Region 2 USDA Forest Service, and Chuck Davis, U.S. -

Mccown's Longspur (Rhynchophanes Mccownii)
McCown’s Longspur (Rhynchophanes mccownii ) A Species Conservation Assessment for The Nebraska Natural Legacy Project Prepared by Melissa J. Panella Nebraska Game and Parks Commission Wildlife Division November 2012 The mission of the Nebraska Natural Legacy Project is to implement a blueprint for conserving Nebraska’s flora, fauna and natural habitats through the proactive, voluntary conservation actions of partners, communities and individuals. Purpose The primary goal in development of at-risk species conservation assessments is to compile biological and ecological information that may assist conservation practitioners in making decisions regarding the conservation of species of interest. The Nebraska Natural Legacy Project recognizes the McCown’s Longspur ( Rhynchophanes mccownii ) as a Tier I at- risk species. Some general management recommendations are made here regarding the McCown’s Longspur (MCLO); however, conservation practitioners will need to use professional judgment to make specific management decisions based on objectives, location, and a multitude of variables. This resource was designed to share available knowledge of MCLO that will aid in the decision-making process or in identifying research needs for the benefit of the species. Species conservation assessments should not be stagnant documents but rather will need to be updated as new scientific information becomes available. The Nebraska Natural Legacy Project focuses efforts in the state’s Biologically Unique Landscapes (BULs), but it is recommended that whenever -

Yukon Wild Buckwheat Eriogonum Flavum Var. Aquilinum
COSEWIC Assessment and Status Report on the Yukon Wild Buckwheat Eriogonum flavum var. aquilinum in Canada SPECIAL CONCERN 2017 COSEWIC status reports are working documents used in assigning the status of wildlife species suspected of being at risk. This report may be cited as follows: COSEWIC. 2017. COSEWIC assessment and status report on the Yukon Wild Buckwheat Eriogonum flavum var. aquilinum in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. x + 32 pp. (http://www.registrelep-sararegistry.gc.ca/default.asp?lang=en&n=24F7211B-1). Production note: COSEWIC would like to acknowledge Allan Harris and Robert Foster for writing the status report on Yukon Wild Buckwheat, Eriogonum flavum var. aquilinum, in Canada, prepared under contract with Environment and Climate Change Canada. This report was overseen and edited by Jana Vamosi, Co-chair of the COSEWIC Vascular Plants Specialist Subcommittee, with significant input from Bruce Bennett, former Co-chair of COSEWIC Vascular Plants and current COSEWIC Vascular Plants Specialist Subcommittee member. For additional copies contact: COSEWIC Secretariat c/o Canadian Wildlife Service Environment and Climate Change Canada Ottawa, ON K1A 0H3 Tel.: 819-938-4125 Fax: 819-938-3984 E-mail: [email protected] http://www.cosewic.gc.ca Également disponible en français sous le titre Ếvaluation et Rapport de situation du COSEPAC sur L’ériogone du Nord (Eriogonum flavum var. aquilinum) au Canada. Cover illustration/photo: Yukon Wild Buckwheat — Photo credit: Allan Harris. Her Majesty the Queen in Right of Canada, 2017. Catalogue No. CW69-14/758-2018E-PDF ISBN 978-0-660-26676-3 COSEWIC Assessment Summary Assessment Summary – November 2017 Common name Yukon Wild Buckwheat Scientific name Eriogonum flavum var. -

Appendix M: Forest Service Biological Evaluation and Management
Final EIS USFS Biological Evaluation Appendix M APPENDIX M—FOREST SERVICE BIOLOGICAL EVALUATION AND MANAGEMENT INDICATOR SPECIES REPORT 1 Final EIS USFS Biological Evaluation Appendix M Forest Service Biological Evaluation for the Greater Sage-Grouse Conservation Effort to Amend the Bridger-Teton and Medicine Bow National Forest Plans and the Thunder Basin National Grassland Management Plan May 13 17, 2015 Prepared by Tim Byer, Wildlife Biologist [email protected] Steve Loose, Wildlife Biologist [email protected] Katharine Haynes, Botanist [email protected] Chris Colt, Wildlife Biologist [email protected] 2 Final EIS USFS Biological Evaluation Appendix M Table of Contents I. INTRODUCTION ................................................................................................................. 11 II. PROJECT HISTORY ........................................................................................................ 12 III. PURPOSE AND NEED ..................................................................................................... 13 IV. DESCRIPTION OF THE ALTERNATIVES .................................................................... 14 Alternative A: No-Action .......................................................................................................... 15 Alternative B: ............................................................................................................................ 15 Alternative C: ........................................................................................................................... -

Pollinator Research Action Plan
POLLINATOR RESEARCH ACTION PLAN Report of the Pollinator Health Task Force MAY 19, 2015 May 19, 2015 On behalf of the Pollinator Health Task Force, we are pleased to transmit the National Strategy to Promote the Health of Honey Bees and Other Pollinators (Strategy). Developed through a collaborative effort across the Executive Branch, this Strategy outlines a comprehensive approach to tackling and reducing the impact of multiple stressors on pollinator health, including pests and pathogens, reduced habitat, lack of nutritional resources, and exposure to pesticides. Building on the current state of the science, and with a renewed emphasis on expanding our understanding of the complex interactions among the various factors impacting pollinator health, the Strategy lays out current and planned Federal actions to achieve the following overarching goals: • Honey Bees: Reduce honey bee colony losses during winter (overwintering mortality) to no more than 15% within 10 years. This goal is informed by the previously released Bee Informed Partnership surveys and the newly established quarterly and annual surveys by the USDA National Agricultural Statistics Service. Based on the robust data anticipated from the national, statistically-based NASS surveys of beekeepers, the Task Force will develop baseline data and additional goal metrics for winter, summer, and total annual colony loss. • Monarch Butterflies: Increase the Eastern population of the monarch butterfly to 225 mil- lion butterflies occupying an area of approximately 15 acres (6 hectares) in the overwintering grounds in Mexico, through domestic/international actions and public-private partnerships, by 2020. • Pollinator Habitat Acreage: Restore or enhance 7 million acres of land for pollinators over the next 5 years through Federal actions and public/private partnerships. -

Coefficients of Conservatism for the Vascular Flora of the Dakotas and Adjacent Grasslands
(200) B565i no. 2001 -1 mt of the Interior al Survey c.1 Coefficients of Conservatism for the Vascular Flora of the Dakotas and Adjacent Grasslands Information and Technology Report USGS/BRD/ITR- 2001 -0001 ~USGS science for a changing world Cover art: Penstemon nitidus courtesy of Dorothy E. Mushet. U.S. Department of the Interior U.S. Geological Survey Coefficients of Conservatism for the Vascular Flora of the Dakotas and Adjacent Grasslands By The Northern Great Plains Floristic Quality Assessment Panel Information and Technology Report USGS/BRD/ITR- 2001 -0001 OCT 1 G 2.001 U.S. Department of the Interior Gale A. Norton, Secretary U.S. Geological Survey Charles G. Groat, Director U.S. Geological Survey, Reston, Virginia: 2001 Any use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the U.S. Government. Cop ies of th is publication are available from the National Technical Information Service, 5285 Port Royal Road, Spring field, Virginia 22161 (1 -800-553-6847 or 703-487 -4650). Copies also are available to registered users from the Defense Technical Information Center, Attn .: Help Desk, 8725 Kingman Road, Suite 0944, Fort Belvoir, Virginia 22060-6218 (1 -800 -225-3842 or 703 -767 - 9050). Suggested citation: The orthem Great Plain Floristic Quality Assessment Panel, 200 I, Coefficient of conservatism for the vascular flora of the Dakotas and adjacent grasslands: U.S. Geological Survey, Biological Resources Divi ion, Lnformation and Technology Report USGS/ BRD/lTR- 200 1-0001, 32 p. Contents Page Ab tract ........... -

Plants Natural Heritage and Natural Legacy Ranks 2018 Nebraska Game and Parks Commission
Species of Greatest Conservation Need Plants Natural Heritage and Natural Legacy Ranks 2018 Nebraska Game and Parks Commission Heritage S1 and /or Legacy Tier 1 Heritage Legacy Heritage Common Name Scientific Name Srank Tier Grank White Baneberry Actaea pachypoda S1 2 G5 Ohio Buckeye Aesculus glabra var. arguta S1S2 2 G5T4?Q Lavender Giant-hyssop Agastache foeniculum S1 2 G5 Purple Giant-hyssop Agastache scrophulariifolia S1 2 G4 Pale False-dandelion Agoseris glauca var. glauca S1 2 G5T5 Spikebent Agrostis exarata var. minor S1 2 G5TNRQ Drummond's Wild Onion Allium drummondii S1 2 G5 Perennial Bursage Ambrosia tomentosa S1 2 G4 Ear-leaf Toothcup Ammannia auriculata S1 2 G5 Dwarf Indigo-bush Amorpha nana S1 2 G5 Prairie Broomweed Amphiachyris dracunculoides S1S2 2 G4G5 Pearly Everlasting Anaphalis margaritacea S1 2 G5 Hudson Bay Anemone Anemone multifida var. multifida S1 2 G5T5 Low Pussytoes Antennaria dimorpha S1 2 G5 Mountain White-camas Anticlea elegans var. elegans S1 2 G5T5 Spikenard Aralia racemosa S1 2 G5 Bearberry Arctostaphylos uva-ursi S1 2 G5 Arrowfeather Three-awn Aristida purpurascens var. purpurascens S1 2 G5T5 Carruth's Sagewort Artemisia carruthii S1 2 G4? Clasping-leaf Milkweed Asclepias amplexicaulis S1 2 G5 Antelope-Horns Asclepias asperula var. decumbens S1 2 G5TNR Field Milk-vetch Astragalus agrestis S1 2 G5 Barr’s Milkvetch Astragalus barrii S1 1 G3 Bodin's Milk-vetch Astragalus bodinii S1 2 G4 Tine-leaf Milk-vetch Astragalus pectinatus S1 2 G5 Short’s Milkvetch Astragalus shortianus S1 1 G4 Gardner's Saltbush Atriplex gardneri var. gardneri S1 2 G5TNR Texas Bergia Bergia texana S1 2 G5 Mountain Birch Betula occidentalis S1 2 G5 Sandberg's Beggar-ticks Bidens connata var. -

U97JON08.Pdf (651.2Kb)
ECOLOGICAL EVALUATION OF THE POTENTIAL ROCK CREEK RESEARCH NATURAL AREA WITHIN THE THUNDER BASIN NATIONAL GRASSLAND, WESTON COUNTY, WYOMING Prepared for Nebraska National Forest, USDA Forest Service By George P. Jones Wyoming Natural Diversity Database The Nature Conservancy 1604 Grand Avenue Laramie, Wyoming 82070 July 16, 1997 ECOLOGICAL EVALUATION OF THE POTENTIAL ROCK CREEK RESEARCH NATURAL AREA WITHIN THE THUNDER BASIN NATIONAL GRASSLAND, WESTON COUNTY, WYOMING Prepared for Nebraska National Forest, USDA Forest Service By George P. Jones Wyoming Natural Diversity Database I, ' The Nature Conservancy 1604 Grand Avenue Laramie, Wyoming 82070 July 16, 1997 TABLE OF CONTENTS INTRODUCTION • • . • • . • . 1 ! Land Management Planning . 1 OBJECTIVES . 1 ' PRINCIPAL DISTINGUISHING FEATURES . • • 2 ' I l__ ' LOCATION • • • • • • • • 2 i I . 2 ' ' Boundary : ~ Area .• . 2 Elevation • • • • • • • • • • 2 Access • . 2 Ecoregion . • . • . • . 3 : \ Maps • . • . • . • . 3 • • 3 : AREA BY COVER TYPES ' I The Vegetation . 3 Area by Type • . 4 PHYSICAL AND CLIMATIC CONDITIONS . 5 Physical Setting . 5 DESCRIPTION OF VALUES . • . • . 5 Vegetation Types . • . • . • 5 Flora . 5 Threatened, Endangered, and Sensitive Plant Species 5 Plant Species Li~t . • . • . • • . 5 Fauna . 8 I Threatened, Endangered, and Sensitive Vertebrates . 8 I ' Geology . 8 'L Lands . 8 SUITABILITY FOR RESEARCH NATURAL AREA SELECTION . 8 Quality . 8 Condition . 9 Viability . 11 Defensibility . • . 12 Degree to Which the Potential RNA Meets Criteria . • • 12 IMPACTS AND POSSIBLE CONFLICTS . 12 Mineral Resources . 13 Grazing . )• . 13 Timber . 13 Watershed Values . 13 Recreation Values . 13 Wildlife and Plant Values . • . 13 Transportation Values . 13 MANAGEMENT CONCERNS 13 REFERENCES 14 Appendix 1. Maps of the potential Rock Creek Research Natural Area. 16 Appendix 2. Photographs from the potential Rock Creek Research Natural Area. -
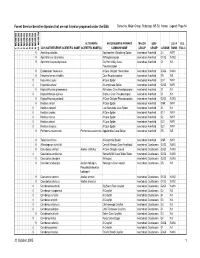
Sensitive Species That Are Not Listed Or Proposed Under the ESA Sorted By: Major Group, Subgroup, NS Sci
Forest Service Sensitive Species that are not listed or proposed under the ESA Sorted by: Major Group, Subgroup, NS Sci. Name; Legend: Page 94 REGION 10 REGION 1 REGION 2 REGION 3 REGION 4 REGION 5 REGION 6 REGION 8 REGION 9 ALTERNATE NATURESERVE PRIMARY MAJOR SUB- U.S. N U.S. 2005 NATURESERVE SCIENTIFIC NAME SCIENTIFIC NAME(S) COMMON NAME GROUP GROUP G RANK RANK ESA C 9 Anahita punctulata Southeastern Wandering Spider Invertebrate Arachnid G4 NNR 9 Apochthonius indianensis A Pseudoscorpion Invertebrate Arachnid G1G2 N1N2 9 Apochthonius paucispinosus Dry Fork Valley Cave Invertebrate Arachnid G1 N1 Pseudoscorpion 9 Erebomaster flavescens A Cave Obligate Harvestman Invertebrate Arachnid G3G4 N3N4 9 Hesperochernes mirabilis Cave Psuedoscorpion Invertebrate Arachnid G5 N5 8 Hypochilus coylei A Cave Spider Invertebrate Arachnid G3? NNR 8 Hypochilus sheari A Lampshade Spider Invertebrate Arachnid G2G3 NNR 9 Kleptochthonius griseomanus An Indiana Cave Pseudoscorpion Invertebrate Arachnid G1 N1 8 Kleptochthonius orpheus Orpheus Cave Pseudoscorpion Invertebrate Arachnid G1 N1 9 Kleptochthonius packardi A Cave Obligate Pseudoscorpion Invertebrate Arachnid G2G3 N2N3 9 Nesticus carteri A Cave Spider Invertebrate Arachnid GNR NNR 8 Nesticus cooperi Lost Nantahala Cave Spider Invertebrate Arachnid G1 N1 8 Nesticus crosbyi A Cave Spider Invertebrate Arachnid G1? NNR 8 Nesticus mimus A Cave Spider Invertebrate Arachnid G2 NNR 8 Nesticus sheari A Cave Spider Invertebrate Arachnid G2? NNR 8 Nesticus silvanus A Cave Spider Invertebrate Arachnid G2? NNR -

2019 Data Report
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science Plant Community Composition and Structure Monitoring at Badlands National Park 2019 Data Report Natural Resource Data Series NPS/BADL/NRDS—2019/1244 ON THIS PAGE The vibrant flowers of Dwarf false indigo, Amorpha nana, were seen at plot PCM_0082 in Badlands National Park this year. Photograph courtesy of the National Park Service ON THE COVER Northern Great Plains Inventory & Monitoring crew members identifying plants at a plot in Badlands National Park. Photograph courtesy of the National Park Service Plant Community Composition and Structure Monitoring at Badlands National Park 2019 Data Report Natural Resource Data Series NPS/BADL/NRDS—2019/1244 Molly B. Davis1 and Daniel J. Swanson2 1National Park Service Northern Great Plains Inventory & Monitoring Network 820 Columbus St. Rapid City, SD 57701 2National Park Service Northern Great Plains Fire Management 26611 U.S. Hwy 385 Hot Springs, SD 57747 November 2019 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Data Series is intended for the timely release of basic data sets and data summaries. Care has been taken to assure accuracy of raw data values, but a thorough analysis and interpretation of the data has not been completed. -

Vegetation Classification and Mapping Project Report
USGS-NPS Vegetation Mapping Program Badlands National Par k USGS-NPS Vegetation Mapping Program Badlands National Park, South Dakota Jim Von Loh ACS Government Solutions Group Dan Cogan ACS Government Solutions Group Don Faber-Langendoen The Nature Conservancy Doug Crawford Bureau of Reclamation Michael J. Pucherelli Bureau of Reclamation November 19, 1999 U.S. Department of the Interior Bureau of Reclamation’s Remote Sensing and GIS Group Denver Federal Center Denver, Colorado The Reclamation Remote Sensing and Geographic Information Group, organized in 1975 provides advice and assistance regarding the application of remote sensing and geographic information system (GIS) technologies to meet the spatial information needs of Bureau of Reclamation and other clients. USGS-NPS Vegetation Mapping Program Badlands National Park TABLE OF CONTENTS LIST OF TABLES...........................................................................................................................IV LIST OF FIGURES.......................................................................................................................... .V LIST OF CONTACTS ..................................................................................................................... VI LIST OF CONTRIBUTORS.............................................................................................................VIII ACKNOWLEDGEMENTS ................................................................................................................ IX EXECUTIVE SUMMARY .........................................................................................................