29Th Annual Molecular Parasitology & Vector Biology Symposium
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Elite Music Productions This Music Guide Represents the Most Requested Songs at Weddings and Parties
Elite Music Productions This Music Guide represents the most requested songs at Weddings and Parties. Please circle songs you like and cross out the ones you don’t. You can also write-in additional requests on the back page! WEDDING SONGS ALL TIME PARTY FAVORITES CEREMONY MUSIC CELEBRATION THE TWIST HERE COMES THE BRIDE WE’RE HAVIN’ A PARTY SHOUT GOOD FEELIN’ HOLIDAY THE WEDDING MARCH IN THE MOOD YMCA FATHER OF THE BRIDE OLD TIME ROCK N ROLL BACK IN TIME INTRODUCTION MUSIC IT TAKES TWO STAYIN ALIVE ST. ELMOS FIRE, A NIGHT TO REMEMBER, RUNAROUND SUE MEN IN BLACK WHAT I LIKE ABOUT YOU RAPPERS DELIGHT GET READY FOR THIS, HERE COMES THE BRIDE BROWN EYED GIRL MAMBO #5 (DISCO VERSION), ROCKY THEME, LOVE & GETTIN’ JIGGY WITH IT LIVIN, LA VIDA LOCA MARRIAGE, JEFFERSONS THEME, BANG BANG EVERYBODY DANCE NOW WE LIKE TO PARTY OH WHAT A NIGHT HOT IN HERE BRIDE WITH FATHER DADDY’S LITTLE GIRL, I LOVED HER FIRST, DADDY’S HANDS, FATHER’S EYES, BUTTERFLY GROUP DANCES KISSES, HAVE I TOLD YOU LATELY, HERO, I’LL ALWAYS LOVE YOU, IF I COULD WRITE A SONG, CHICKEN DANCE ALLEY CAT CONGA LINE ELECTRIC SLIDE MORE, ONE IN A MILLION, THROUGH THE HANDS UP HOKEY POKEY YEARS, TIME IN A BOTTLE, UNFORGETTABLE, NEW YORK NEW YORK WALTZ WIND BENEATH MY WINGS, YOU LIGHT UP MY TANGO YMCA LIFE, YOU’RE THE INSPIRATION LINDY MAMBO #5BAD GROOM WITH MOTHER CUPID SHUFFLE STROLL YOU RAISE ME UP, TIMES OF MY LIFE, SPECIAL DOLLAR WINE DANCE MACERENA ANGEL, HOLDING BACK THE YEARS, YOU AND CHA CHA SLIDE COTTON EYED JOE ME AGAINST THE WORLD, CLOSE TO YOU, MR. -

The Ride in the Saddle
Copyright Page This book was automatically created by FLAG on April 19th, 2012, based on content retrieved from http://www.fanfiction.net/s/5838394/. The content in this book is copyrighted by Aylah50 or their authorised agent(s). All rights are reserved except where explicitly stated otherwise. This story was first published on March 23rd, 2010, and was last updated on March 25th, 2011. Any and all feedback is greatly appreciated - please email any bugs, problems, feature requests etc. to [email protected]. Table of Contents Summary 1. Chapter 1: Lightning Crashes 2. Chapter 2: I Wanna Be Sedated 3. Chapter 3: Alive 4. Chapter 4: Take On Me 5. Chapter 5: Sweet Child of Mine 6. Chapter 6: Everything You Want 7. Chapter 7: Wicked Garden 8. Chapter 8: Selling the Drama 9. Chapter 9: Now That We've Found Love 10. Chapter 10: Breaking the Habit 11. Chapter 11: The World I Know 12. Chapter 12: Holiday 13. Chapter 13: Pour Some Sugar On Me 14. Chapter 14: Everybody Hurts 15. Chapter 15: This I Promise You 16. Chapter 16: What a Girl Wants 17. Chapter 17: The Reason 18. Chapter 18: Possession 19. Chapter 19: Ghost of Me 20. Chapter 20: Call Your Name 21. Chapter 21: Home 22. Chapter 22 I Still Haven't Found 23. Chapter 23: Broken 24. Chapter 24: In the Air Tonight 25. Chapter 25: It's Not My Time 26. Chapter 26: I Alone 27. Chapter 27: Don't Wanna Miss a Thing 28. Chapter 28: Life is a Highway - 3 - 29. -

The Role of Genetic Variation in Predisposition to Alcohol-Related Chronic Pancreatitis
The Role of Genetic Variation in Predisposition to Alcohol-related Chronic Pancreatitis Thesis submitted in accordance with the requirements of the University of Liverpool for the degree of Doctor in Philosophy by Marianne Lucy Johnstone April 2015 The Role of Genetic Variation in Predisposition to Alcohol-related Chronic Pancreatitis 2015 Abstract Background Chronic pancreatitis (CP) is a disease of fibrosis of the pancreas for which alcohol is the main causative agent. However, only a small proportion of alcoholics develop chronic pancreatitis. Genetic polymorphism may affect pancreatitis risk. Aim To determine the factors required to classify a chronic pancreatic population and identify genetic variations that may explain why only some alcoholics develop chronic pancreatitis. Methods The most appropriate method of diagnosing CP was assessed using a systematic review. Genetics of different populations of alcohol-related chronic pancreatitics (ACP) were explored using four different techniques: genome-wide association study (GWAS); custom arrays; PCR of variable nucleotide tandem repeats (VNTR) and next generation sequencing (NGS) of selected genes. Results EUS and sMR were identified as giving the overall best sensitivity and specificity for diagnosing CP. GWAS revealed two associations with CP (identified and replicated) at PRSS1-PRSS2_rs10273639 (OR 0.73, 95% CI 0.68-0.79) and X-linked CLDN2_rs12688220 (OR 1.39, 1.28-1.49) and the association was more pronounced in the ACP group (OR 0.56, 0.48-0.64)and OR 2.11, 1.84-2.42). The previously identified VNTR in CEL was shown to have a lower frequency of the normal repeat in ACP than alcoholic liver disease (ALD; OR 0.61, 0.41-0.93). -

Genes in Eyecare Geneseyedoc 3 W.M
Genes in Eyecare geneseyedoc 3 W.M. Lyle and T.D. Williams 15 Mar 04 This information has been gathered from several sources; however, the principal source is V. A. McKusick’s Mendelian Inheritance in Man on CD-ROM. Baltimore, Johns Hopkins University Press, 1998. Other sources include McKusick’s, Mendelian Inheritance in Man. Catalogs of Human Genes and Genetic Disorders. Baltimore. Johns Hopkins University Press 1998 (12th edition). http://www.ncbi.nlm.nih.gov/Omim See also S.P.Daiger, L.S. Sullivan, and B.J.F. Rossiter Ret Net http://www.sph.uth.tmc.edu/Retnet disease.htm/. Also E.I. Traboulsi’s, Genetic Diseases of the Eye, New York, Oxford University Press, 1998. And Genetics in Primary Eyecare and Clinical Medicine by M.R. Seashore and R.S.Wappner, Appleton and Lange 1996. M. Ridley’s book Genome published in 2000 by Perennial provides additional information. Ridley estimates that we have 60,000 to 80,000 genes. See also R.M. Henig’s book The Monk in the Garden: The Lost and Found Genius of Gregor Mendel, published by Houghton Mifflin in 2001 which tells about the Father of Genetics. The 3rd edition of F. H. Roy’s book Ocular Syndromes and Systemic Diseases published by Lippincott Williams & Wilkins in 2002 facilitates differential diagnosis. Additional information is provided in D. Pavan-Langston’s Manual of Ocular Diagnosis and Therapy (5th edition) published by Lippincott Williams & Wilkins in 2002. M.A. Foote wrote Basic Human Genetics for Medical Writers in the AMWA Journal 2002;17:7-17. A compilation such as this might suggest that one gene = one disease. -

Dottorato Di Ricerca the Effect of Finasteride
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by UniCA Eprints Università degli Studi di Cagliari DOTTORATO DI RICERCA Scuola di Dottorato in Neuroscienze e Scienze Morfologiche Corso di Dottorato in Neuroscienze Ciclo XXIII THE EFFECT OF FINASTERIDE IN TOURETTE SYNDROME: RESULTS OF A CLINICAL TRIAL Settore scientifico disciplinari di afferenza BIO/14 Presentata da: Silvia Paba Coordinatore Dottorato Prof.ssa Alessandra Concas Tutor Dott.ssa Paola Devoto Esame finale anno accademico 2009 - 2010 1. INTRODUCTION 1 1.1 General characteristics of steroid 5α-reductase 2 1.2 S5αR inhibitors 15 1.3 S5αR inhibitors as putative therapeutic agents for some neuropsychiatric disorders. 23 2. AIMS OF THE STUDY 37 3. METHODS 38 3.1 Subjects 38 3.2 Procedures 39 3.3 Data analysis 40 4. RESULTS 41 4.1 Description of sample 41 4.2 Dosing, range and compliance 41 4.3 Effects of finasteride on TS and tic severity 44 4.4 Effects of finasteride on obsessive compulsive symptoms 46 4.5 Adverse effects 47 5. DISCUSSION 48 6. CONCLUSION 52 REFERENCES 54 1. INTRODUCTION The enzyme steroid 5α reductase (S5αR) catalyzes the conversion of Δ4-3-ketosteroid precursors - such as testosterone, progesterone and androstenedione - into their 5α- reduced metabolites. Although the current nomenclature assigns five enzymes to the S5αR family, only the types 1 and 2 appear to play an important role in steroidogenesis, mediating an overlapping set of reactions, albeit with distinct chemical characteristics and anatomical distribution. The discovery that the 5α-reduced metabolite of testosterone, 5α-dihydrotestosterone (DHT), is the most potent androgen and stimulates prostatic growth led to the development of S5αR inhibitors with high efficacy and tolerability. -

A Journey Through the Inner World of Def Leppard
THE MANY FACES OF DEF LEPPARD A JOURNEY THROUGH THE INNER WORLD OF DEF LEPPARD 2LP 7798093712896 RELEASE: 04-12-2020 With more than 65 million albums sold worldwide and two prestigious Diamond Awards to their credit, Def Leppard with its current line-up –Joe Elliott (vocals), Phil Collen (guitar), Rick Savage LIMITED EDITION (bass), Vivian Campbell (guitar) and Rick Allen (drums)— continue to be one of the most important GATEFOLD TRANSPARENT forces in rock music.n many ways, ORANGE/BLUE MARBLED 180 GR. VINYL Def Leppard were the definitive hard rock band of the ‘80s. There were groups that rocked harder, but few captured the spirit of the times quite as well. Emerging in the late ‘70s as part of the New Wave of British Heavy Metal, Def Leppard gained a following outside of that scene by toning down their heavy riffs and emphasizing melody. In The Many Faces of Def Leppard, we are going to dig deep into the lesser known work of their members, collaborations, side projects and of course, their hit-filled catalog. LP1 - THE MANY FACES OF DEF LEPPARD LP2 - THE SONGS Side A Side A ALL JOIN HANDS - ROADHOUSE FEAT. PETE WILLIS & FRANK NOON POUR SOME SUGAR ON ME - LEE THOMPSON, JOHNNY DEE, RICHARD KENDRICK, I WILL BE THERE - GOMAGOG FEAT. PETE WILLIS & JANICK GERS MARKO PUKKILA & HOWIE SIMON DOIN’ FINE - CARMINE APPICE FEAT. VIVIAN CAMPBELL HYSTERIA - DANIEL FLORES ON A LONELY ROAD - THE FROG & THE RICK VITO BAND LET IT GO - WICKED GARDEN FEAT. JOE ELLIOTT ROCK ROCK TIL YOU DROP - CHRIS POLAND, JOE VIERS & RICHARD KENDRICK OVER MY DEAD BODY - MAN RAZE FEAT. -

Def Leppard “Hysteria” By: Kyle Harman
Def Leppard “Hysteria” By: Kyle Harman Hysteria would be a fitting name for the album thought Def Leppard drummer Rick Allen, following his 1984 injury from a horrific car accident that resulted in the loss of his left arm, Hysteria was the perfect title to describe the worldwide media coverage that followed the band between the years of 1984 and 1987. The album was the last to feature the late Steve Clark and his role of lead guitarist due to an overdose in the year of 1992. In the very prime years of glam metal in the late 80’s at the dawn of grunge genre about to begin, Def Leppard delivered the bands bestselling album to date with selling 25 million copies worldwide and 12 million in the US alone. The three year recording process of Hysteria was painstaking with the accident of drummer Rick Allen, and with producers coming and going left the band with the perfect example of sometimes patience truly does pay off. A unique recording process took place where each member recorded their own recordings at separate times, allowing each member to focus on their own performance on the album. The original intent of the album was to be a rock version of Michael Jackson’s Thriller, with the desire of having every song off the album as a possible hit single. The end result of the album was impressive to say the least with 7 singles released off the single album alone. The album recording sessions were painstaking at times, “Animal” consisted of 2 and a half years to produce its final product, where “Pour Some Sugar On Me” was finished within two weeks. -
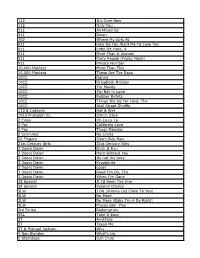
112 It's Over Now 112 Only You 311 All Mixed up 311 Down
112 It's Over Now 112 Only You 311 All Mixed Up 311 Down 702 Where My Girls At 911 How Do You Want Me To Love You 911 Little Bit More, A 911 More Than A Woman 911 Party People (Friday Night) 911 Private Number 10,000 Maniacs More Than This 10,000 Maniacs These Are The Days 10CC Donna 10CC Dreadlock Holiday 10CC I'm Mandy 10CC I'm Not In Love 10CC Rubber Bullets 10CC Things We Do For Love, The 10CC Wall Street Shuffle 112 & Ludacris Hot & Wet 1910 Fruitgum Co. Simon Says 2 Evisa Oh La La La 2 Pac California Love 2 Pac Thugz Mansion 2 Unlimited No Limits 20 Fingers Short Dick Man 21st Century Girls 21st Century Girls 3 Doors Down Duck & Run 3 Doors Down Here Without You 3 Doors Down Its not my time 3 Doors Down Kryptonite 3 Doors Down Loser 3 Doors Down Road I'm On, The 3 Doors Down When I'm Gone 38 Special If I'd Been The One 38 Special Second Chance 3LW I Do (Wanna Get Close To You) 3LW No More 3LW No More (Baby I'm A Do Right) 3LW Playas Gon' Play 3rd Strike Redemption 3SL Take It Easy 3T Anything 3T Tease Me 3T & Michael Jackson Why 4 Non Blondes What's Up 5 Stairsteps Ooh Child 50 Cent Disco Inferno 50 Cent If I Can't 50 Cent In Da Club 50 Cent In Da Club 50 Cent P.I.M.P. (Radio Version) 50 Cent Wanksta 50 Cent & Eminem Patiently Waiting 50 Cent & Nate Dogg 21 Questions 5th Dimension Aquarius_Let the sunshine inB 5th Dimension One less Bell to answer 5th Dimension Stoned Soul Picnic 5th Dimension Up Up & Away 5th Dimension Wedding Blue Bells 5th Dimension, The Last Night I Didn't Get To Sleep At All 69 Boys Tootsie Roll 8 Stops 7 Question -

1. Rush - Tom Sawyer 2
1. Rush - Tom Sawyer 2. Black Sabbath - Iron Man 3. Def Leppard - Photograph 4. Guns N Roses - November Rain 5. Led Zeppelin - Stairway to Heaven 6. AC/DC - Highway To Hell 7. Aerosmith - Sweet Emotion 8. Metallica - Enter Sandman 9. Nirvana - All Apologies 10. Black Sabbath - War Pigs 11. Guns N Roses - Paradise City 12. Tom Petty - Refugee 13. Journey - Wheel In The Sky 14. Led Zeppelin - Kashmir 15. Jimi Hendrix - All Along The Watchtower 16. ZZ Top - La Grange 17. Pink Floyd - Another Brick In The Wall 18. Black Sabbath - Paranoid 19. Soundgarden - Black Hole Sun 20. Motley Crue - Home Sweet Home 21. Derek & The Dominoes - Layla 22. Rush - The Spirit Of Radio 23. Led Zeppelin - Rock And Roll 24. The Who - Baba O Riley 25. Foreigner - Hot Blooded 26. Lynyrd Skynyrd - Freebird 27. ZZ Top - Cheap Sunglasses 28. Pearl Jam - Alive 29. Dio - Rainbow In The Dark 30. Deep Purple - Smoke On The Water 31. Nirvana - In Bloom 32. Jimi Hendrix - Voodoo Child (Slight Return) 33. Billy Squier - In The Dark 34. Pink Floyd - Wish You Were Here 35. Rush - Working Man 36. Led Zeppelin - Whole Lotta Love 37. Black Sabbath - NIB 38. Foghat - Slow Ride 39. Def Leppard - Armageddon It 40. Boston - More Than A Feeling 41. AC/DC - Dirty Deeds Done Dirt Cheap 42. Ozzy Osbourne - Crazy Train 43. Aerosmith - Walk This Way 44. Led Zeppelin - Dazed & Confused 45. Def Leppard - Rock Of Ages 46. Ozzy Osbourne - No More Tears 47. Eric Clapton - Cocaine 48. Ted Nugent - Stranglehold 49. Aerosmith - Dream On 50. Van Halen - Eruption / You Really Got Me 51. -

Cell-Specific Proteomic Analysis in Caenorhabditis Elegans
Supporting Information Appendix (265 Pages) for Cell-Specific Proteomic Analysis in Caenorhabditis elegans Authors: Kai P. Yueta, Meenakshi K. Domab, c, John T. Ngoa, 2, Michael J. Sweredoskid, Robert L. J. Grahamd, 3, Annie Moradiand, Sonja Hessd, Erin M. Schumane, Paul W. Sternbergb,c and David A. Tirrella,1 Author Affiliations: aDivision of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California, United States of America bDivision of Biology and Biological Engineering, California Institute of Technology, Pasadena, California, United States of America cHoward Hughes Medical Institute, California Institute of Technology, Pasadena, California, United States of America dProteome Exploration Laboratory, Beckman Institute, California Institute of Technology, Pasadena, California, United States of America eMax Planck Institute for Brain Research, Frankfurt am Main, Germany 1To whom correspondence may be addressed. 2Current Address: Department of Pharmacology, University of California, San Diego, La Jolla, California, United States of America 3Current Address: Faculty of Medical and Human Sciences, University of Manchester, Manchester, United Kingdom Supporting Information Materials and Methods - Adenosine Triphosphate (ATP)-Pyrophosphate (PPi) Exchange Assay - Chloroform/Methanol Precipitation - Enrichment of p-Azido-L-Phenylalanine-Labeled Proteins - Fluorescence Microscopy of Live C. elegans - Fluorescence Microscopy of p-Azido-L-Phenylalanine-Labeled Proteins in Fixed C. elegans - In-Gel Fluorescence -

Def Leppard Headline Grand Opening Concert
DEF LEPPARD TO HEADLINE GRAND OPENING CONCERT FOR HARD ROCK HOTEL & CASINO SACRAMENTO AT FIRE MOUNTAIN DEF LEPPARD WITH DON FELDER AND LAST IN LINE SATURDAY, NOVEMBER 2ND TOYOTA AMPHITHEATRE Tickets On Sale Friday, September 13 at 10am at LiveNation.com Hard Rock Hotel & Casino Sacramento at Fire Mountain Set to Open on October 30 SACRAMENTO, Calif. – Sept. 6, 2019 – With great anticipation, Hard Rock Hotel & Casino Sacramento at Fire Mountain announces the property will open its doors the evening of Wednesday, October 30, with grand opening celebrations occurring all weekend. The official opening will kick off with a world-famous Hard Rock guitar smash; and in true Hard Rock fashion, Def Leppard will perform on Saturday, November 2 at Toyota Amphitheatre with special guests Don Felder and Last in Line to commemorate the opening. On Sunday, November 3 guests can experience some of the most exciting marketing promotions and events in the marketplace. “We’ve been looking forward to opening our doors since we broke ground in July 2018,” said Mark Birtha, president of Hard Rock Hotel & Casino Sacramento at Fire Mountain. “Hard Rock is passionate about providing world-class service in amazingly designed venues to create authentic experiences that rock. We’re excited for our guests to experience all of that and many more surprises here in Northern California. This milestone has been made possible through our relationships with local businesses and contractors, the dedication of 1,300 team members, and the vision, commitment and hard work of the Enterprise Rancheria Tribe.” Do you want to get Rocked! On Saturday, November 2, BRITISH ICONS Def Leppard will rock the stage as the Hard Rock’s grand opening performance at Toyota Amphitheatre next door. -

European Patent Office U.S. Patent and Trademark Office
EUROPEAN PATENT OFFICE U.S. PATENT AND TRADEMARK OFFICE CPC NOTICE OF CHANGES 89 DATE: JULY 1, 2015 PROJECT RP0098 The following classification changes will be effected by this Notice of Changes: Action Subclass Group(s) Symbols deleted: C12Y 101/01063 C12Y 101/01128 C12Y 101/01161 C12Y 102/0104 C12Y 102/03011 C12Y 103/01004 C12Y 103/0103 C12Y 103/01052 C12Y 103/99007 C12Y 103/9901 C12Y 103/99013 C12Y 103/99021 C12Y 105/99001 C12Y 105/99002 C12Y 113/11013 C12Y 113/12012 C12Y 114/15002 C12Y 114/99028 C12Y 204/01119 C12Y 402/01052 C12Y 402/01058 C12Y 402/0106 C12Y 402/01061 C12Y 601/01025 C12Y 603/02027 Symbols newly created: C12Y 101/01318 C12Y 101/01319 C12Y 101/0132 C12Y 101/01321 C12Y 101/01322 C12Y 101/01323 C12Y 101/01324 C12Y 101/01325 C12Y 101/01326 C12Y 101/01327 C12Y 101/01328 C12Y 101/01329 C12Y 101/0133 C12Y 101/01331 C12Y 101/01332 C12Y 101/01333 CPC Form – v.4 CPC NOTICE OF CHANGES 89 DATE: JULY 1, 2015 PROJECT RP0098 Action Subclass Group(s) C12Y 101/01334 C12Y 101/01335 C12Y 101/01336 C12Y 101/01337 C12Y 101/01338 C12Y 101/01339 C12Y 101/0134 C12Y 101/01341 C12Y 101/01342 C12Y 101/03043 C12Y 101/03044 C12Y 101/98003 C12Y 101/99038 C12Y 102/01083 C12Y 102/01084 C12Y 102/01085 C12Y 102/01086 C12Y 103/01092 C12Y 103/01093 C12Y 103/01094 C12Y 103/01095 C12Y 103/01096 C12Y 103/01097 C12Y 103/0701 C12Y 103/08003 C12Y 103/08004 C12Y 103/08005 C12Y 103/08006 C12Y 103/08007 C12Y 103/08008 C12Y 103/08009 C12Y 103/99032 C12Y 104/01023 C12Y 104/01024 C12Y 104/03024 C12Y 105/01043 C12Y 105/01044 C12Y 105/01045 C12Y 105/03019 C12Y 105/0302