Materials Studio Overview
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Introducing ONETEP: Linear-Scaling Density Functional Simulations on Parallel Computers Chris-Kriton Skylaris,A) Peter D
THE JOURNAL OF CHEMICAL PHYSICS 122, 084119 ͑2005͒ Introducing ONETEP: Linear-scaling density functional simulations on parallel computers Chris-Kriton Skylaris,a) Peter D. Haynes, Arash A. Mostofi, and Mike C. Payne Theory of Condensed Matter, Cavendish Laboratory, Madingley Road, Cambridge CB3 0HE, United Kingdom ͑Received 29 September 2004; accepted 4 November 2004; published online 23 February 2005͒ We present ONETEP ͑order-N electronic total energy package͒, a density functional program for parallel computers whose computational cost scales linearly with the number of atoms and the number of processors. ONETEP is based on our reformulation of the plane wave pseudopotential method which exploits the electronic localization that is inherent in systems with a nonvanishing band gap. We summarize the theoretical developments that enable the direct optimization of strictly localized quantities expressed in terms of a delocalized plane wave basis. These same localized quantities lead us to a physical way of dividing the computational effort among many processors to allow calculations to be performed efficiently on parallel supercomputers. We show with examples that ONETEP achieves excellent speedups with increasing numbers of processors and confirm that the time taken by ONETEP as a function of increasing number of atoms for a given number of processors is indeed linear. What distinguishes our approach is that the localization is achieved in a controlled and mathematically consistent manner so that ONETEP obtains the same accuracy as conventional cubic-scaling plane wave approaches and offers fast and stable convergence. We expect that calculations with ONETEP have the potential to provide quantitative theoretical predictions for problems involving thousands of atoms such as those often encountered in nanoscience and biophysics. -

Natural Bond Orbital Analysis in the ONETEP Code: Applications to Large Protein Systems Louis P
WWW.C-CHEM.ORG FULL PAPER Natural Bond Orbital Analysis in the ONETEP Code: Applications to Large Protein Systems Louis P. Lee,*[a] Daniel J. Cole,[a] Mike C. Payne,[a] and Chris-Kriton Skylaris[b] First principles electronic structure calculations are typically Generalized Wannier Functions of ONETEP to natural atomic performed in terms of molecular orbitals (or bands), providing a orbitals, NBO analysis can be performed within a localized straightforward theoretical avenue for approximations of region in such a way that ensures the results are identical to an increasing sophistication, but do not usually provide any analysis on the full system. We demonstrate the capabilities of qualitative chemical information about the system. We can this approach by performing illustrative studies of large derive such information via post-processing using natural bond proteins—namely, investigating changes in charge transfer orbital (NBO) analysis, which produces a chemical picture of between the heme group of myoglobin and its ligands with bonding in terms of localized Lewis-type bond and lone pair increasing system size and between a protein and its explicit orbitals that we can use to understand molecular structure and solvent, estimating the contribution of electronic delocalization interactions. We present NBO analysis of large-scale calculations to the stabilization of hydrogen bonds in the binding pocket of with the ONETEP linear-scaling density functional theory package, a drug-receptor complex, and observing, in situ, the n ! p* which we have interfaced with the NBO 5 analysis program. In hyperconjugative interactions between carbonyl groups that ONETEP calculations involving thousands of atoms, one is typically stabilize protein backbones. -
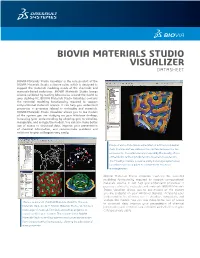
Biovia Materials Studio Visualizer Datasheet
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. -

Download Author Version (PDF)
PCCP Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/pccp Page 1 of 11 PhysicalPlease Chemistry do not adjust Chemical margins Physics PCCP PAPER Effect of nanosize on surface properties of NiO nanoparticles for adsorption of Quinolin-65 ab a a Received 00th January 20xx, Nedal N. Marei, Nashaat N. Nassar* and Gerardo Vitale Accepted 00th January 20xx Using Quinolin-65 (Q-65) as a model-adsorbing compound for polar heavy hydrocarbons, the nanosize effect of NiO Manuscript DOI: 10.1039/x0xx00000x nanoparticles on adsorption of Q-65 was investigated. Different-sized NiO nanoparticles with sizes between 5 and 80 nm were prepared by controlled thermal dehydroxylation of Ni(OH)2. -

Supporting Information
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2015 Supporting Information A Single Crystalline Porphyrinic Titanium Metal−Organic Framework Shuai Yuana†, Tian-Fu Liua†, Dawei Fenga, Jian Tiana, Kecheng Wanga, Junsheng Qina, a a a a b Qiang Zhang , Ying-Pin Chen , Mathieu Bosch , Lanfang Zou , Simon J. Teat, Scott J. c a Dalgarno and Hong-Cai Zhou * a Department of Chemistry, Texas A&M University, College Station, Texas 77842-3012, USA b Advanced Light Source, Lawrence Berkeley National Laboratory Berkeley, CA 94720, USA c Institute of Chemical Sciences, Heriot-Watt University Riccarton, Edinburgh EH14 4AS, U.K. † Equal contribution to this work *To whom correspondence should be addressed. Email: [email protected] Tel: +1 (979) 845-4034; Fax: +1 (979) 845-1595 S1 Contents S1. Ligand Synthesis..............................................................................................................3 S2. Syntheses of PCN-22.......................................................................................................5 S3. X-ray Crystallography .....................................................................................................6 S4. Topological Analyses ......................................................................................................9 S5. N2 Sorption Isotherm .....................................................................................................10 S6. Simulation of the Accessible Surface Area ...................................................................12 -

PDF Hosted at the Radboud Repository of the Radboud University Nijmegen
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/19078 Please be advised that this information was generated on 2021-09-23 and may be subject to change. Computational Chemistry Metho ds Applications to Racemate Resolution and Radical Cation Chemistry ISBN Computational Chemistry Metho ds Applications to Racemate Resolution and Radical Cation Chemistry een wetenschapp elijkeproeve op het gebied van de Natuurwetenschapp en Wiskunde en Informatica Pro efschrift ter verkrijging van de graad van do ctor aan de KatholiekeUniversiteit Nijmegen volgens b esluit van het College van Decanen in het op enbaar te verdedigen op dinsdag januari des namiddags om uur precies do or Gijsb ert Schaftenaar geb oren op augustus te Harderwijk Promotores Prof dr ir A van der Avoird Prof dr E Vlieg Copromotor Prof dr RJ Meier Leden manuscriptcommissie Prof dr G Vriend Prof dr RA de Gro ot Dr ir PES Wormer The research rep orted in this thesis was nancially supp orted by the Dutch Or ganization for the Advancement of Science NWO and DSM Contents Preface Intro duction Intro duction Chirality Metho ds for obtaining pure enantiomers Racemate Resolution via diastereomeric salt formation Rationalization of diastereomeric salt formation Computational metho ds for mo deling the lattice energy Molecular Mechanics Quantum Chemical -

Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins
Article Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins Marcus Purse 1, Grace Edmund 1, Stephen Hall 1, Brendan Howlin 1,* , Ian Hamerton 2 and Stephen Till 3 1 Department of Chemistry, Faculty of Engineering and Physical Sciences, University of Surrey, Guilford, Surrey GU2 7XH, UK; [email protected] (M.P.); [email protected] (G.E.); [email protected] (S.H.) 2 Bristol Composites Institute (ACCIS), Department of Aerospace Engineering, School of Civil, Aerospace, and Mechanical Engineering, University of Bristol, Bristol BS8 1TR, UK; [email protected] 3 Defence Science and Technology Laboratory, Porton Down, Salisbury SP4 0JQ, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-1483-686-248 Received: 6 March 2019; Accepted: 23 March 2019; Published: 28 March 2019 Abstract: The thermal decomposition of polyphenolic resins was studied by reactive molecular dynamics (RMD) simulation at elevated temperatures. Atomistic models of the polyphenolic resins to be used in the RMD were constructed using an automatic method which calls routines from the software package Materials Studio. In order to validate the models, simulated densities and heat capacities were compared with experimental values. The most suitable combination of force field and thermostat for this system was the Forcite force field with the Nosé–Hoover thermostat, which gave values of heat capacity closest to those of the experimental values. Simulated densities approached a final density of 1.05–1.08 g/cm3 which compared favorably with the experimental values of 1.16–1.21 g/cm3 for phenol-formaldehyde resins. -

Dmol Guide to Select a Dmol3 Task 1
DMOL3 GUIDE MATERIALS STUDIO 8.0 Copyright Notice ©2014 Dassault Systèmes. All rights reserved. 3DEXPERIENCE, the Compass icon and the 3DS logo, CATIA, SOLIDWORKS, ENOVIA, DELMIA, SIMULIA, GEOVIA, EXALEAD, 3D VIA, BIOVIA and NETVIBES are commercial trademarks or registered trademarks of Dassault Systèmes or its subsidiaries in the U.S. and/or other countries. All other trademarks are owned by their respective owners. Use of any Dassault Systèmes or its subsidiaries trademarks is subject to their express written approval. Acknowledgments and References To print photographs or files of computational results (figures and/or data) obtained using BIOVIA software, acknowledge the source in an appropriate format. For example: "Computational results obtained using software programs from Dassault Systèmes Biovia Corp.. The ab initio calculations were performed with the DMol3 program, and graphical displays generated with Materials Studio." BIOVIA may grant permission to republish or reprint its copyrighted materials. Requests should be submitted to BIOVIA Support, either through electronic mail to [email protected], or in writing to: BIOVIA Support 5005 Wateridge Vista Drive, San Diego, CA 92121 USA Contents DMol3 1 Setting up a molecular dynamics calculation20 Introduction 1 Choosing an ensemble 21 Further Information 1 Defining the time step 21 Tasks in DMol3 2 Defining the thermostat control 21 Energy 3 Constraints during dynamics 21 Setting up the calculation 3 Setting up a transition state calculation 22 Dynamics 4 Which method to use? -

Density Functional Theory (DFT)
Herramientas mecano-cuánticas basadas en DFT para el estudio de moléculas y materiales en Materials Studio 7.0 Javier Ramos Biophysics of Macromolecular Systems group (BIOPHYM) Departamento de Física Macromolecular Instituto de Estructura de la Materia – CSIC [email protected] Webinar, 26 de Junio 2014 Anteriores webinars Como conseguir los videos y las presentaciones de anteriores webminars: Linkedin: Grupo de Química Computacional http://www.linkedin.com/groups/Química-computacional-7487634 Índice Density Functional Theory (DFT) The Jacob’s ladder DFT modules in Maretials Studio DMOL3, CASTEP and ONETEP XC functionals Basis functions Interfaces in Materials Studio Tasks Properties Example: n-butane conformations Density Functional Theory (DFT) DFT is built around the premise that the energy of an electronic system can be defined in terms of its electron probability density (ρ). (Hohenberg-Kohn Theorem) E 0 [ 0 ] Te [ 0 ] E ne [ 0 ] E ee [ 0 ] (easy) Kinetic Energy for ????? noninteracting (r )v (r ) dr electrons(easy) 1 E[]()()[]1 r r d r d r E e e2 1 2 1 2 X C r12 Classic Term(Coulomb) Non-classic Kohn-Sham orbitals Exchange & By minimizing the total energy functional applying the variational principle it is Correlation possible to get the SCF equations (Kohn-Sham) The Jacob’s Ladder Accurate form of XC potential Meta GGA Empirical (Fitting to Non-Empirical Generalized Gradient Approx. atomic properties) (physics rules) Local Density Approximation DFT modules in Materials Studio DMol3: Combine computational speed with the accuracy of quantum mechanical methods to predict materials properties reliably and quickly CASTEP: CASTEP offers simulation capabilities not found elsewhere, such as accurate prediction of phonon spectra, dielectric constants, and optical properties. -

Compact Orbitals Enable Low-Cost Linear-Scaling Ab Initio Molecular Dynamics for Weakly-Interacting Systems Hayden Scheiber,1, A) Yifei Shi,1 and Rustam Z
Compact orbitals enable low-cost linear-scaling ab initio molecular dynamics for weakly-interacting systems Hayden Scheiber,1, a) Yifei Shi,1 and Rustam Z. Khaliullin1, b) Department of Chemistry, McGill University, 801 Sherbrooke St. West, Montreal, QC H3A 0B8, Canada Today, ab initio molecular dynamics (AIMD) relies on the locality of one-electron density matrices to achieve linear growth of computation time with systems size, crucial in large-scale simulations. While Kohn-Sham orbitals strictly localized within predefined radii can offer substantial computational advantages over density matrices, such compact orbitals are not used in AIMD because a compact representation of the electronic ground state is difficult to find. Here, a robust method for maintaining compact orbitals close to the ground state is coupled with a modified Langevin integrator to produce stable nuclear dynamics for molecular and ionic systems. This eliminates a density matrix optimization and enables first orbital-only linear-scaling AIMD. An application to liquid water demonstrates that low computational overhead of the new method makes it ideal for routine medium-scale simulations while its linear-scaling complexity allows to extend first- principle studies of molecular systems to completely new physical phenomena on previously inaccessible length scales. Since the unification of molecular dynamics and den- LS methods restrict their use in dynamical simulations sity functional theory (DFT)1, ab initio molecular dy- to very short time scales, systems of low dimensions, namics (AIMD) has become an important tool to study and low-quality minimal basis sets6,18–20. On typical processes in molecules and materials. Unfortunately, the length and time scales required in practical and accurate computational cost of the conventional Kohn-Sham (KS) AIMD simulations, LS DFT still cannot compete with DFT grows cubically with the number of atoms, which the straightforward low-cost cubically-scaling KS DFT. -

Quantum Chemistry (QC) on Gpus Feb
Quantum Chemistry (QC) on GPUs Feb. 2, 2017 Overview of Life & Material Accelerated Apps MD: All key codes are GPU-accelerated QC: All key codes are ported or optimizing Great multi-GPU performance Focus on using GPU-accelerated math libraries, OpenACC directives Focus on dense (up to 16) GPU nodes &/or large # of GPU nodes GPU-accelerated and available today: ACEMD*, AMBER (PMEMD)*, BAND, CHARMM, DESMOND, ESPResso, ABINIT, ACES III, ADF, BigDFT, CP2K, GAMESS, GAMESS- Folding@Home, GPUgrid.net, GROMACS, HALMD, HOOMD-Blue*, UK, GPAW, LATTE, LSDalton, LSMS, MOLCAS, MOPAC2012, LAMMPS, Lattice Microbes*, mdcore, MELD, miniMD, NAMD, NWChem, OCTOPUS*, PEtot, QUICK, Q-Chem, QMCPack, OpenMM, PolyFTS, SOP-GPU* & more Quantum Espresso/PWscf, QUICK, TeraChem* Active GPU acceleration projects: CASTEP, GAMESS, Gaussian, ONETEP, Quantum Supercharger Library*, VASP & more green* = application where >90% of the workload is on GPU 2 MD vs. QC on GPUs “Classical” Molecular Dynamics Quantum Chemistry (MO, PW, DFT, Semi-Emp) Simulates positions of atoms over time; Calculates electronic properties; chemical-biological or ground state, excited states, spectral properties, chemical-material behaviors making/breaking bonds, physical properties Forces calculated from simple empirical formulas Forces derived from electron wave function (bond rearrangement generally forbidden) (bond rearrangement OK, e.g., bond energies) Up to millions of atoms Up to a few thousand atoms Solvent included without difficulty Generally in a vacuum but if needed, solvent treated classically -

Introduction to DFT and the Plane-Wave Pseudopotential Method
Introduction to DFT and the plane-wave pseudopotential method Keith Refson STFC Rutherford Appleton Laboratory Chilton, Didcot, OXON OX11 0QX 23 Apr 2014 Parallel Materials Modelling Packages @ EPCC 1 / 55 Introduction Synopsis Motivation Some ab initio codes Quantum-mechanical approaches Density Functional Theory Electronic Structure of Condensed Phases Total-energy calculations Introduction Basis sets Plane-waves and Pseudopotentials How to solve the equations Parallel Materials Modelling Packages @ EPCC 2 / 55 Synopsis Introduction A guided tour inside the “black box” of ab-initio simulation. Synopsis • Motivation • The rise of quantum-mechanical simulations. Some ab initio codes Wavefunction-based theory • Density-functional theory (DFT) Quantum-mechanical • approaches Quantum theory in periodic boundaries • Plane-wave and other basis sets Density Functional • Theory SCF solvers • Molecular Dynamics Electronic Structure of Condensed Phases Recommended Reading and Further Study Total-energy calculations • Basis sets Jorge Kohanoff Electronic Structure Calculations for Solids and Molecules, Plane-waves and Theory and Computational Methods, Cambridge, ISBN-13: 9780521815918 Pseudopotentials • Dominik Marx, J¨urg Hutter Ab Initio Molecular Dynamics: Basic Theory and How to solve the Advanced Methods Cambridge University Press, ISBN: 0521898633 equations • Richard M. Martin Electronic Structure: Basic Theory and Practical Methods: Basic Theory and Practical Density Functional Approaches Vol 1 Cambridge University Press, ISBN: 0521782856