Regateiro Et Al Supplementary Figure 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Epigenetic Control of Mammalian Centromere Protein Binding: Does DNA Methylation Have a Role?
Journal of Cell Science 109, 2199-2206 (1996) 2199 Printed in Great Britain © The Company of Biologists Limited 1996 JCS3386 Epigenetic control of mammalian centromere protein binding: does DNA methylation have a role? Arthur R. Mitchell*, Peter Jeppesen, Linda Nicol†, Harris Morrison and David Kipling MRC Human Genetics Unit, Western General Hospital, Crewe Road, Edinburgh EH4 2XU, UK *Author for correspondence (internet [email protected]) †Present address: MRC Reproductive Biology Unit, Edinburgh, UK SUMMARY Chromosome 1 of the inbred mouse strain DBA/2 has a block of minor satellite DNA sequences on chromosome 1. polymorphism associated with the minor satellite DNA at The binding of the CENP-E protein does not appear to be its centromere. The more terminal block of satellite DNA affected by demethylation of the minor satellite sequences. sequences on this chromosome acts as the centromere as We present a model to explain these observations. This shown by the binding of CREST ACA serum, anti-CENP- model may also indicate the mechanism by which the B and anti-CENP-E polyclonal sera. Demethylation of the CENP-B protein recognises specific sites within the arrays minor satellite DNA sequences accomplished by growing of minor satellite DNA on mouse chromosomes. cells in the presence of the drug 5-aza-2′-deoxycytidine results in a redistribution of the CENP-B protein. This protein now binds to an enlarged area on the more terminal Key words: Centromere satellite DNA, Demethylation, Centromere block and in addition it now binds to the more internal antibody INTRODUCTION A common feature of many mammalian pericentromeric domains is that they contain families of repetitive DNA The centromere of mammalian chromosomes is recognised at sequences (Singer, 1982). -

Chapter 1 - Introduction
Chapter 1 - Introduction 1 1.1 Epigenetic modifications and gene silencing The fact that a multicellular organism is able to undergo cellular differentiation, even though every cell contains the same genetic material, indicates the existence of an extra layer of phenotype-determining factors. Cellular differentiation is associated with the development of different patterns of gene expression in individual cells or groups of cells, and the way in which these patterns are established involves epigenetics. Epigenetics refers to the process by which transcriptional states are changed in a relatively permanent way in the absence of DNA mutation. Epigenetic gene silencing or activation results from modifications to the gene in question either by changing the methylation state of the DNA or by modifying the proteins that package the DNA. There is increasing evidence that in some cases RNA molecules are also involved. Epigenetic modifications are generally erased and reset between generations in order to provide the necessary pluripotent starting state from which to begin development in the next generation. 1.1.1 DNA methylation DNA methylation is the best studied epigenetic modification. In mammals DNA methylation is always associated with cytosine residues, generally occurring at CpG dinucleotides in a symmetrical fashion, where the cytosines on both strands are methylated. Methylation of cytosine involves the transfer of a methyl group from an S-adenosyl-L-methionine to the 5-position of cytosine, resulting in the modified base 5-methylcytosine (Jeltsch, 2002) (Figure 1.1). Estimates of methylation levels at CpGs in mammals range from 50-90% (Gruenbaum et al., 1981; Jeltsch, 2002). DNA 2 hypermethylation at promoters usually correlates with transcriptional repression and gene silencing. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Sexual Dimorphism in Brain Transcriptomes of Amami Spiny Rats (Tokudaia Osimensis): a Rodent Species Where Males Lack the Y Chromosome Madison T
Ortega et al. BMC Genomics (2019) 20:87 https://doi.org/10.1186/s12864-019-5426-6 RESEARCHARTICLE Open Access Sexual dimorphism in brain transcriptomes of Amami spiny rats (Tokudaia osimensis): a rodent species where males lack the Y chromosome Madison T. Ortega1,2, Nathan J. Bivens3, Takamichi Jogahara4, Asato Kuroiwa5, Scott A. Givan1,6,7,8 and Cheryl S. Rosenfeld1,2,8,9* Abstract Background: Brain sexual differentiation is sculpted by precise coordination of steroid hormones during development. Programming of several brain regions in males depends upon aromatase conversion of testosterone to estrogen. However, it is not clear the direct contribution that Y chromosome associated genes, especially sex- determining region Y (Sry), might exert on brain sexual differentiation in therian mammals. Two species of spiny rats: Amami spiny rat (Tokudaia osimensis) and Tokunoshima spiny rat (T. tokunoshimensis) lack a Y chromosome/Sry, and these individuals possess an XO chromosome system in both sexes. Both Tokudaia species are highly endangered. To assess the neural transcriptome profile in male and female Amami spiny rats, RNA was isolated from brain samples of adult male and female spiny rats that had died accidentally and used for RNAseq analyses. Results: RNAseq analyses confirmed that several genes and individual transcripts were differentially expressed between males and females. In males, seminal vesicle secretory protein 5 (Svs5) and cytochrome P450 1B1 (Cyp1b1) genes were significantly elevated compared to females, whereas serine (or cysteine) peptidase inhibitor, clade A, member 3 N (Serpina3n) was upregulated in females. Many individual transcripts elevated in males included those encoding for zinc finger proteins, e.g. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Complex Network-Driven View of Genomic Mechanisms Underlying Parkinson’S Disease: Analyses in Dorsal Motor Vagal Nucleus, Locus Coeruleus, and Substantia Nigra
Hindawi Publishing Corporation BioMed Research International Volume 2014, Article ID 543673, 16 pages http://dx.doi.org/10.1155/2014/543673 Research Article Complex Network-Driven View of Genomic Mechanisms Underlying Parkinson’s Disease: Analyses in Dorsal Motor Vagal Nucleus, Locus Coeruleus, and Substantia Nigra Beatriz Raposo Corradini,1 Priscila Iamashita,1 Edilaine Tampellini,2,3 José Marcelo Farfel,3,4 Lea Tenenholz Grinberg,2,5,6 and Carlos Alberto Moreira-Filho1 1 Department of Pediatrics, Faculdade de Medicina da USP (FMUSP), Avenida Dr. Eneas´ Carvalho Aguiar 647, 5 Andar, 05403-900 Sao˜ Paulo, SP, Brazil 2 Brazilian Aging Brain Study Group (BEHEEC), LIM 22, FMUSP, 01246-903 Sao˜ Paulo, SP, Brazil 3 Hospital Israelita Albert Einstein, 05652-900 Sao˜ Paulo, SP, Brazil 4 Division of Geriatrics, FMUSP, 01246-903 Sao˜ Paulo, SP, Brazil 5 Department of Pathology, FMUSP, 01246-903 Sao˜ Paulo, SP, Brazil 6 Department of Neurology and Pathology, University of California, San Francisco, CA 94143, USA Correspondence should be addressed to Carlos Alberto Moreira-Filho; [email protected] Received 28 May 2014; Accepted 15 September 2014; Published 26 November 2014 Academic Editor: Meike Kasten Copyright © 2014 Beatriz Raposo Corradini et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Parkinson’s disease (PD)—classically characterized by severe loss of dopaminergic neurons in the substantia nigra pars compacta— has a caudal-rostral progression, beginning in the dorsal motor vagal nucleus and, in a less extent, in the olfactory system, progressing to the midbrain and eventually to the basal forebrain and the neocortex. -

Satellite DNA at the Centromere Is Dispensable for Segregation Fidelity
G C A T T A C G G C A T genes Brief Report Satellite DNA at the Centromere Is Dispensable for Segregation Fidelity Annalisa Roberti, Mirella Bensi, Alice Mazzagatti, Francesca M. Piras, Solomon G. Nergadze , Elena Giulotto * and Elena Raimondi * Department of Biology and Biotechnology “L. Spallanzani”, University of Pavia, Via Ferrata 1, 27100 Pavia, Italy; [email protected] (A.R.); [email protected] (M.B.); [email protected] (A.M.); [email protected] (F.M.P.); [email protected] (S.G.N.) * Correspondence: [email protected] (E.G.); [email protected] (E.R.) Received: 7 June 2019; Accepted: 19 June 2019; Published: 20 June 2019 Abstract: The typical vertebrate centromeres contain long stretches of highly repeated DNA sequences (satellite DNA). We previously demonstrated that the karyotypes of the species belonging to the genus Equus are characterized by the presence of satellite-free and satellite-based centromeres and represent a unique biological model for the study of centromere organization and behavior. Using horse primary fibroblasts cultured in vitro, we compared the segregation fidelity of chromosome 11, whose centromere is satellite-free, with that of chromosome 13, which has similar size and a centromere containing long stretches of satellite DNA. The mitotic stability of the two chromosomes was compared under normal conditions and under mitotic stress induced by the spindle inhibitor, nocodazole. Two independent molecular-cytogenetic approaches were used—the interphase aneuploidy analysis and the cytokinesis-block micronucleus assay. Both assays were coupled to fluorescence in situ hybridization with chromosome specific probes in order to identify chromosome 11 and chromosome 13, respectively. -

Identification and Characterization of Satellite Dnas in Two-Toed Sloths Of
www.nature.com/scientificreports OPEN Identifcation and characterization of satellite DNAs in two‑toed sloths of the genus Choloepus (Megalonychidae, Xenarthra) Radarane Santos Sena1, Pedro Heringer1, Mirela Pelizaro Valeri1, Valéria Socorro Pereira2, Gustavo C. S. Kuhn1 & Marta Svartman1* Choloepus, the only extant genus of the Megalonychidae family, is composed of two living species of two‑toed sloths: Choloepus didactylus and C. hofmanni. In this work, we identifed and characterized the main satellite DNAs (satDNAs) in the sequenced genomes of these two species. SATCHO1, the most abundant satDNA in both species, is composed of 117 bp tandem repeat sequences. The second most abundant satDNA, SATCHO2, is composed of ~ 2292 bp tandem repeats. Fluorescence in situ hybridization in C. hofmanni revealed that both satDNAs are located in the centromeric regions of all chromosomes, except the X. In fact, these satDNAs present some centromeric characteristics in their sequences, such as dyad symmetries predicted to form secondary structures. PCR experiments indicated the presence of SATCHO1 sequences in two other Xenarthra species: the tree‑toed sloth Bradypus variegatus and the anteater Myrmecophaga tridactyla. Nevertheless, SATCHO1 is present as large tandem arrays only in Choloepus species, thus likely representing a satDNA exclusively in this genus. Our results reveal interesting features of the satDNA landscape in Choloepus species with the potential to aid future phylogenetic studies in Xenarthra and mammalian genomes in general. A signifcant part of eukaryotic genomes, ~ 30% in some plants to more than 50% in some insects and mammals, is composed of tandemly organized highly repetitive sequences, known as satellite DNAs (satDNAs) (reviewed in Ref.1). -

1 SUPPLEMENTAL DATA Figure S1. Poly I:C Induces IFN-Β Expression
SUPPLEMENTAL DATA Figure S1. Poly I:C induces IFN-β expression and signaling. Fibroblasts were incubated in media with or without Poly I:C for 24 h. RNA was isolated and processed for microarray analysis. Genes showing >2-fold up- or down-regulation compared to control fibroblasts were analyzed using Ingenuity Pathway Analysis Software (Red color, up-regulation; Green color, down-regulation). The transcripts with known gene identifiers (HUGO gene symbols) were entered into the Ingenuity Pathways Knowledge Base IPA 4.0. Each gene identifier mapped in the Ingenuity Pathways Knowledge Base was termed as a focus gene, which was overlaid into a global molecular network established from the information in the Ingenuity Pathways Knowledge Base. Each network contained a maximum of 35 focus genes. 1 Figure S2. The overlap of genes regulated by Poly I:C and by IFN. Bioinformatics analysis was conducted to generate a list of 2003 genes showing >2 fold up or down- regulation in fibroblasts treated with Poly I:C for 24 h. The overlap of this gene set with the 117 skin gene IFN Core Signature comprised of datasets of skin cells stimulated by IFN (Wong et al, 2012) was generated using Microsoft Excel. 2 Symbol Description polyIC 24h IFN 24h CXCL10 chemokine (C-X-C motif) ligand 10 129 7.14 CCL5 chemokine (C-C motif) ligand 5 118 1.12 CCL5 chemokine (C-C motif) ligand 5 115 1.01 OASL 2'-5'-oligoadenylate synthetase-like 83.3 9.52 CCL8 chemokine (C-C motif) ligand 8 78.5 3.25 IDO1 indoleamine 2,3-dioxygenase 1 76.3 3.5 IFI27 interferon, alpha-inducible -

CENP-B Dynamics at Centromeres Is Regulated by a Sumoylation
bioRxiv preprint doi: https://doi.org/10.1101/245597; this version posted January 9, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 CENP-B dynamics at centromeres is regulated by a SUMOylation/ubiquitination and 2 proteasomal-dependent degradation mechanism involving the SUMO-targeted ubiquitin E3 3 ligase RNF4 4 Jhony El Maalouf 1,, Pascale Texier 1,4, Indri Erliandri 1,4, Camille Cohen 1, Armelle Corpet 1, Frédéric 5 Catez 2, Chris Boutell 3, Patrick Lomonte 1,* 6 7 1. Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5310, INSERM U 1217, LabEx DEVweCAN, 8 Institut NeuroMyoGène (INMG), team Chromatin Assembly, Nuclear Domains, Virus. F-69100, Lyon, 9 France 10 2. Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5286, INSERM U 1052, Centre de 11 Recherche en Cancérologie de Lyon. F-69000, Lyon, France 12 3. MRC-University of Glasgow Centre for Virus Research, Glasgow G61 1QH, Scotland (UK) 13 4. Contributed equally to this work. 14 15 * Corresponding author: [email protected] 16 17 18 Running title: SUMO & ubiquitin-dependent CENP-B dynamics 19 20 Word count: 6743 21 22 23 24 The English of parts of this document has been checked by at least two professional editors, both native 25 speakers of English. For a certificate, please see: 26 27 http://www.textcheck.com/certificate/gh7EcX 28 29 30 1 bioRxiv preprint doi: https://doi.org/10.1101/245597; this version posted January 9, 2018. -

PRODUCT SPECIFICATION Product Datasheet
Product Datasheet QPrEST PRODUCT SPECIFICATION Product Name QPrEST ZN112 Mass Spectrometry Protein Standard Product Number QPrEST38525 Protein Name Zinc finger protein 112 Uniprot ID Q9UJU3 Gene ZNF112 Product Description Stable isotope-labeled standard for absolute protein quantification of Zinc finger protein 112. Lys (13C and 15N) and Arg (13C and 15N) metabolically labeled recombinant human protein fragment. Application Absolute protein quantification using mass spectrometry Sequence (excluding EREEKLLMVETETPRDGCSGRKNQQKMESIQEVTVSYFSPKELSSRQTWQ fusion tag) QSAGGLIRCQDFLKVFQGKNSQLQEQGNSLGQVWAGIPVQISEDKNYIFT HIGNGSNYIKSQGYPSWRAHHSWRKMYLKESHNYQCRCQQI Theoretical MW 34308 Da including N-terminal His6ABP fusion tag Fusion Tag A purification and quantification tag (QTag) consisting of a hexahistidine sequence followed by an Albumin Binding Protein (ABP) domain derived from Streptococcal Protein G. Expression Host Escherichia coli LysA ArgA BL21(DE3) Purification IMAC purification Purity >90% as determined by Bioanalyzer Protein 230 Purity Assay Isotopic Incorporation >99% Concentration >5 μM after reconstitution in 100 μl H20 Concentration Concentration determined by LC-MS/MS using a highly pure amino acid analyzed internal Determination reference (QTag), CV ≤10%. Amount >0.5 nmol per vial, two vials supplied. Formulation Lyophilized in 100 mM Tris-HCl 5% Trehalose, pH 8.0 Instructions for Spin vial before opening. Add 100 μL ultrapure H2O to the vial. Vortex thoroughly and spin Reconstitution down. For further dilution, see Application Protocol. Shipping Shipped at ambient temperature Storage Lyophilized product shall be stored at -20°C. See COA for expiry date. Reconstituted product can be stored at -20°C for up to 4 weeks. Avoid repeated freeze-thaw cycles. Notes For research use only Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. -
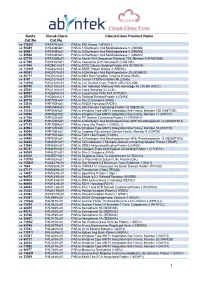
1 Santa Cat.No Cloud-Clone Cat.No. Cloud-Clone Product Name
Santa Cloud -Clone Cloud -Clone Product Name Cat.No Cat.No. sc -130253 PAS477Hu01 PAB to RIO Kinase 1 (RIOK1) sc -50485 PAS446Ra01 PAB to A Disintegrin And Metalloprotease 5 (ADAM5) sc -50487 PAS445Ra01 PAB to A Disintegrin And Metalloprotease 6 (ADAM6) sc -25588 PAS444Ra01 PAB to A Disintegrin And Metalloprotease 1 (ADAM1) sc -87719 PAR758Mu01 PAB to Family With Sequence Similarity 135, Member B (FAM135B) sc -67296 PAR493Hu01 PAB to Coenzyme Q10 Homolog B (COQ10B) sc -67048 PAQ981Hu01 PAB to S100 Calcium Binding Protein A15 (S100A15) sc -130269 PAQ342Hu01 PAB to SRSF Protein Kinase 3 (SRPK3) sc -98582 PAQ207Hu01 PAB to A Disintegrin And Metalloprotease 20 (ADAM20) sc -20711 PAQ164Hu01 PAB to BMX Non Receptor Tyrosine Kinase (BMX) sc -9147 PAQ127Hu01 PAB to Cluster Of Differentiation 8b (CD8b) sc -130184 PAQ124Hu01 PAB to Cell Division Cycle Protein 25B (CDC25B) sc -98790 PAQ118Hu01 PAB to Cell Adhesion Molecule With Homology To L1CAM (CHL1) sc -25361 PAQ116Hu01 PAB to Clock Homolog (CLOCK) sc -98937 PAQ089Hu01 PAB to Cytochrome P450 3A7 (CYP3A7) sc -25519 PAQ088Hu01 PAB to Dickkopf Related Protein 4 (DKK4) sc -28778 PAP797Hu01 PAB to Pim-2 Oncogene (PIM2) sc -33626 PAP750Hu01 PAB to RAD51 Homolog (RAD51) sc -8333 PAP694Mu01 PAB to SH2 Domain Containing Protein 1A (SH2D1A) sc -25524 PAP553Hu01 PAB to Wingless Type MMTV Integration Site Family, Member 10B (WNT10B) sc -50360 PAP552Hu01 PAB to Wingless Type MMTV Integration Site Family, Member 11 (WNT11) sc -87368 PAP332Hu01 PAB to PR Domain Containing Protein 14 (PRDM14) sc -25583 PAP226Hu01