UNIVERSITY of CALIFORNIA Los Angeles Improving the Detection
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Download (739Kb)
VIEWS & REVIEWS Arterial ischemic stroke in HIV Defining and classifying etiology for research studies Laura A. Benjamin, ABSTRACT MRCP, PhD HIV infection, and potentially its treatment, increases the risk of an arterial ischemic stroke. Mul- Alan Bryer, FCN (SA), tiple etiologies and lack of clear case definitions inhibit progress in this field. Several etiologies, PhD many treatable, are relevant to HIV-related stroke. To fully understand the mechanisms and the Sebastian Lucas, FRCP, terminology used, a robust classification algorithm to help ascribe the various etiologies is FRCPath needed. This consensus paper considers the strengths and limitations of current case definitions Alan Stanley, FCN (SA) in the context of HIV infection. The case definitions for the major etiologies in HIV-related strokes Theresa J. Allain, FRCP, were refined (e.g., varicella zoster vasculopathy and antiphospholipid syndrome) and in some in- PhD stances new case definitions were described (e.g., HIV-associated vasculopathy). These case def- Elizabeth Joekes, FRCR initions provided a framework for an algorithm to help assign a final diagnosis, and help classify Hedley Emsley, FRCP, the subtypes of HIV etiology in ischemic stroke. Neurol Neuroimmunol Neuroinflamm 2016;3:e254; PhD doi: 10.1212/NXI.0000000000000254 Ian Turnbull, FRCR Colin Downey, MIBMS GLOSSARY Cheng-Hock Toh, FRCP, ACL 5 anticardiolipin antibodies; anti-b2GP1 5 anti–b2-glycoprotein I; APS 5 antiphospholipid syndrome; HSV 5 herpes PhD simplex virus; IgG 5 immunoglobulin G; LA 5 lupus anticoagulant; RPR 5 rapid plasma reagin; SVD 5 small vessel disease; 5 5 5 Kevin Brown, FRCPath, TB tuberculosis; TOAST Trial of Org 10172 in Acute Stroke Treatment; TTP thrombotic thrombocytopenic purpura; VDRL 5 Venereal Disease Research Laboratory; VZV 5 varicella zoster virus. -

Herpesviruses
J Clin Pathol: first published as 10.1136/jcp.32.9.859 on 1 September 1979. Downloaded from Journal of Clinical Pathology, 1979, 32, 859-881 Herpesviruses MORAG C. TIMBURY' AND ELIZABETH EDMOND2 From the 'Department of Bacteriology, Royal Infirmary, Glasgow and the 2Regional Virus Laboratory, City Hospital, Greenbank Drive, Edinburgh, UK Herpesviruses are ubiquitous in both human and tion (Plummer et al., 1970) or microneutralisation animal populations (Plummer, 1967). The four tests (Pauls and Dowdle, 1967) are often used for human herpesviruses are herpes simplex (HSV), this, but differentiating the two types of virus today varicella-zoster (VZ), cytomegalovirus (CMV), and can probably be done more easily by biochemical Epstein-Barr (EBV) viruses, and all exhibit the prop- methods. Thus the DNA of the viruses can be dis- erty, rare among human pathogenic viruses, of tinguished by restriction enzyme analysis (Skare et remaining latent within the body after primary infec- al., 1975). Similarly, many of the virus polypeptides tion. Latent virus persists for many years-probably produced in infected cells by the two types of virus throughout life-and in some patients reactivates to can be distinguished by polyacrylamide gel electro- cause secondary or recurrent infections. Human phoresis (Courtney and Powell, 1975). herpesviruses can almost be regarded as part of the commensal flora, and certainly HSV is present in LABORATORY DIAGNOSIS the saliva of healthy people from time to time HSV-1 infections are most rapidly diagnosed by (Douglas and Couch, 1970). The viruses exhibit a isolation of the virus in cell cultures such as BHK21 remarkably successful parasitism since the upset to or RK1 3 cells (Grist et al., 1979). -

New York Chapter American College of Physicians Annual
New York Chapter American College of Physicians Annual Scientific Meeting Poster Presentations Saturday, October 12, 2019 Westchester Hilton Hotel 699 Westchester Avenue Rye Brook, NY New York Chapter American College of Physicians Annual Scientific Meeting Medical Student Clinical Vignette 1 Medical Student Clinical Vignette Adina Amin Medical Student Jessy Epstein, Miguel Lacayo, Emmanuel Morakinyo Touro College of Osteopathic Medicine A Series of Unfortunate Events - A Rare Presentation of Thoracic Outlet Syndrome Venous thoracic outlet syndrome, formerly known as Paget-Schroetter Syndrome, is a condition characterized by spontaneous deep vein thrombosis of the upper extremity. It is a very rare syndrome resulting from anatomical abnormalities of the thoracic outlet, causing thrombosis of the deep veins draining the upper extremity. This disease is also called “effort thrombosis― because of increased association with vigorous and repetitive upper extremity activities. Symptoms include severe upper extremity pain and swelling after strenuous activity. A 31-year-old female with a history of vascular thoracic outlet syndrome, two prior thrombectomies, and right first rib resection presented with symptoms of loss of blood sensation, dull pain in the area, and sharp pain when coughing/sneezing. When the patient had her first blood clot, physical exam was notable for swelling, venous distension, and skin discoloration. The patient had her first thrombectomy in her right upper extremity a couple weeks after the first clot was discovered. Thrombolysis with TPA was initiated, and percutaneous mechanical thrombectomy with angioplasty of the axillary and subclavian veins was performed. Almost immediately after the thrombectomy, the patient had a rethrombosis which was confirmed by ultrasound. -

Antibiotic Use for Sepsis in Neonates and Children: 2016 Evidence Update
Antibiotic Use for Sepsis in Neonates and Children: 2016 Evidence Update Aline Fuchsa, Julia Bielickia,b, Shrey Mathurb, Mike Sharlandb, Johannes N. Van Den Ankera,c a Paediatric Pharmacology and Pharmacometrics, University Children's Hospital Basel, Basel, Switzerland b Paediatric Infectious Disease Research Group, Institute for Infection and Immunity, St George's University of London, London, United Kingdom c Division of Clinical Pharmacology, Children’s National Health System, Washington, DC, USA WHO-Reviews 1 TABLE OF CONTENTS 1. INTRODUCTION ............................................................................................................................... 3 1.1. Aims ......................................................................................................................................... 3 1.2. Background ............................................................................................................................. 3 1.2.1. Definition and diagnosis ................................................................................................. 3 Neonatal Sepsis ............................................................................................................................... 3 Paediatric Sepsis ............................................................................................................................. 4 Community versus hospital acquired sepsis .................................................................................. 5 1.2.2. Microbiology .................................................................................................................. -

The Pangenome Diversity, Dynamics and Evolution of Genomes the Pangenome Hervé Tettelin • Duccio Medini Editors
Hervé Tettelin Duccio Medini Editors The Pangenome Diversity, Dynamics and Evolution of Genomes The Pangenome Hervé Tettelin • Duccio Medini Editors The Pangenome Diversity, Dynamics and Evolution of Genomes Editors Hervé Tettelin Duccio Medini Department of Microbiology and GSK Vaccines R&D Immunology, Institute for Genome Siena, Italy Sciences University of Maryland School of Medicine Baltimore, Maryland, USA ISBN 978-3-030-38280-3 ISBN 978-3-030-38281-0 (eBook) https://doi.org/10.1007/978-3-030-38281-0 This book is an open access publication. © The Editor(s) (if applicable) and The Author(s) 2020. Open Access This book is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence and indicate if changes were made. The images or other third party material in this book are included in the book’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the book’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. -

A Compendium of Transfusion Practice Guidelines Edition 4.0 January 2021
A Compendium of Transfusion Practice Guidelines Edition 4.0 January 2021 Table of Contents Introduction Red Blood Cells Platelets Low Titer Group O Whole Blood Plasma Cryoprecipitated AHF Testing Serices Therapeutic Apheresis Blood Component Modifications Hospital Transfusion Committee Patient Blood Management Appendices Introduction Transfusion of blood products is one of the most common medical procedures, used for patients with a wide range of medical conditions to improve tissue oxygenation, achieve hemostasis, and/or fight infections. Moreover, transfusion therapy enables many complex procedures, such as organ transplantation, cardiac and other surgeries, and stem cell transplantation. Yet the curriculum of academic medical and nursing programs provide limited exposure and training towards helping providers to understand the attributes of the different types of blood products, appreciate the risks of transfusion, and raise awareness of the accumulating body of evidence that is helping to refine our understanding of clinical indications for each type of transfusion. While the approach to transfusion medicine has historically been based on personal experience, local practice, expert opinion, and consensus conference recommendations, the availability of hemovigilance data that document the adverse effects of transfusion, randomized controlled trials (RCTs) demonstrating both the benefits and risks of transfusion, and growing debates regarding alternate therapies provide a good foundation to develop evidence-based resources to aid in transfusion care of today and the future. There is a growing belief that transfusion therapy, like many other types of drug therapies, can be tailored or personalized to address specific patient and disease contexts. One example is in the care of the actively hemorrhaging patient, particularly in the prehospital time frame. -

Prospects for Effective and Scalable Community-Based Approaches to Improve Reproductive, Maternal, Newborn and Child Health (RMNCH)
Prospects for Effective and Scalable Community-Based Approaches to Improve Reproductive, Maternal, Newborn and Child Health (RMNCH) A Summary of Experiences from the Maternal and Child Health Integrated Program (MCHIP) and the Child Survival and Health Grants Program (CSHGP) and a Review of the Evidence June 2014 Authors: Henry B. Perry Jim Ricca Karen LeBan Melanie Morrow Copyright © 2014 by Jhpiego Corporation. All rights reserved. Suggested citation: Perry HB, Ricca J, LeBan K. and Morrow M. 2014. Prospects for Effective and Scalable Community-Based Approaches to Improve Reproductive, Maternal, Newborn and Child Health (RMNCH): A Summary of Experiences from the Maternal and Child Health Integrated Program (MCHIP) and the Child Survival and Health Grants Program (CSHGP) and a Review of the Evidence. Jhpiego. Baltimore, MD. The Maternal and Child Health Integrated Program (MCHIP) is the USAID Bureau for Global Health’s flagship maternal, neonatal and child health (MNCH) program. MCHIP supports programming in maternal, newborn and child health, immunization, family planning, malaria, nutrition, and HIV/AIDS, and strongly encourages opportunities for integration. Cross-cutting technical areas include water, sanitation, hygiene, urban health and health systems strengthening. This report was made possible by the generous support of the American people through the United States Agency for International Development (USAID), under the terms of the Leader with Associates Cooperative Agreement GHS-A-00-08-00002-00. The contents are the responsibility of the Maternal and Child Health Integrated Program (MCHIP) and do not necessarily reflect the views of USAID or the United States Government. Table of Contents Table of Contents .................................................................................................................................... i Abbreviations ......................................................................................................................................... -

Consultation Document for Case Definitions
Consultation Document for Case Definitions Adverse Events of Special Interest and Adverse Events Following Immunization during COVID-19 Vaccine Introduction 0 Consultation Document for Case Definitions Adverse Events of Special Interest and Adverse Events Following Immunization during COVID-19 Vaccine Introduction Washington, D.C., 2021 Consultation Document for Case Definitions: Adverse Events of Special Interest and Adverse Events Following Immunization during COVID-19 Vaccine Introduction PAHO/HSS/MT/COVID-19/21-0006 © Pan American Health Organization, 2021 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO license (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this license, this work may be copied, redistributed, and adapted for non-commercial purposes, provided the new work is issued using the same or equivalent Creative Commons license and it is appropriately cited. In any use of this work, there should be no suggestion that the Pan American Health Organization (PAHO) endorses any specific organization, product, or service. Use of the PAHO logo is not permitted. All reasonable precautions have been taken by PAHO to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either expressed or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall PAHO be liable for damages arising from its use. Disclaimer. This document was prepared with support from the European Union through the project “Working Together to Fight Antimicrobial Resistance”. -

Epstein-Barr Virus
CHAPTER 10 Epstein-Barr Virus Alfred S. Evans and James C. Niederman 1. Introduction 2. Historical Background Epstein-Barr virus (EBV), a member of the herpes In 1889, Emil Pfeiffer of Wiesbaden, Germany, group of viruses, is the cause of heterophile-positive described a condition called Driisenfieber (glandular infectious mononucleosis, of most heterophile-neg fever), characterized by fever, adenopathy, mild ative cases, and of occasional cases of tonsillitis and sore throat, and in severe cases enlargement of the pharyngitis in childhood. Rarely, it may involve the liver and spleen. (63, 126) Since Pfeiffer's original de liver or central nervous system as primary mani scription, as well as Filatov's, (53,54) a Russian, in festations. This virus is strongly implicated as hav 1892, antedated by some 30-50 years recognition of ing a causal relationship to African Burkitt lym the hematological changes and the heterophile an phoma and to nasopharyngeal cancer. High antibody tibody, it is uncertain whether these were true in titers are also present in 30-40% of cases of Hodg fectious mononucleosis. However, his description kin's disease, in some patients with sarcoidosis, and of this febrile syndrome in older children and young in systemic lupus erythematosus. adults seems best to fit this diagnosis. There is little This chapter will deal with the epidemiology of doubt about the classic description of the disease EBV infections and the epidemiology of infectious made by Sprunt and Evans, (139) from Johns Hop mononucleosis. Infectious mononucleosis can be kins, in 1920. They described the disorder in young defined as an acute febrile illness involving children adults as we now know it, named the disease "in and young adults characterized clinically by sore fectious mononucleosis," and reported in detail the throat and lymphadenopathy, hematologically by hematological changes. -

Volume - Ix Issue - L Mar / Apr 2012
VOLUME - IX ISSUE - L MAR / APR 2012 The Disease Diagnosis segment of this issue comprehends for you a disease that has many synonyms - it is simply known as infectious mononucleosis or as EBV infectious mononucleosis, Pfeiffer's disease, Filatov's disease and sometimes colloquially as the kissing disease from its oral transmission or simply as mono in North America and as glandular fever in other English-speaking countries) is an infectious, widespread viral disease caused by the Epstein–Barr virus (EBV), one type of herpes virus, to which more than 90% of adults have been exposed. Occasionally, the symptoms can recur at a later period. Most people are 1 Editorial exposed to the virus as children, when the disease produces no noticeable or only flu-like Disease symptoms. In developing countries, people are exposed to the virus in early childhood more 2 often than in developed countries. As a result, the disease in its observable form is more Diagnosis common in developed countries. It is most common among adolescents and young adults. The Trouble disease is omnipresent the world over. 5 Shooting Especially in adolescents and young adults, the disease is characterized by fever, sore throat and fatigue, along with several other possible signs and symptoms. It is primarily diagnosed by 6 Bouquet observation of symptoms, but suspicion can be confirmed by several diagnostic tests. The syndrome was described as an infectious process by Nil Filatov in 1887 and 7 Interpretation independently by Emil Pfeiffer in 1889. It evokes a very strong reaction from the reticuloendothelial system and the body throws up what are known as Downey & McKinley cells (atypical lymphocytes) that are not of neoplastic 8 Tulip News but as bad looking and can be mistaken for neoplastic cells. -
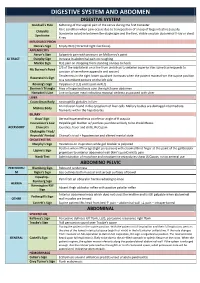
Digestive System and Abdomen
DIGESTIVE SYSTEM AND ABDOMEN DIGESTIVE SYSTEM Goodsall's Rule Softening of the vaginal part of the cervix during the first trimester Rare condition when pain occurs due to transposition of a loop of large intestine (usually Chilaiditi transverse colon) in between the diaphragm and the liver, visible on plain abdominal X-ray or chest Syndrome X-ray INTUSSUSCEPTION Dance's Sign Empty RLQ (retracted right iliac fossa) APPENDICITIS Aaron's Sign Epigastric pain with pressure on McBurney's point GI TRACT Dunphy Sign Increase in abdominal pain on coughing Markle Sign RLQ pain on dropping from standing on toes to heels 2/3 of the way lateral on a line from umbilicus to anterior superior iliac spine (corresponds to Mc Burney’s Point junction of vermiform appendix and cecum) Tenderness in the right lower quadrant increases when the patient moves from the supine position Rosenstein's Sign to a recumbent posture on the left side Rovsing's Sign Palpation of LLQ elicits pain in RLQ Sherren's Triangle Area of hyperaesthesia over the right lower abdomen Hampton's Line Line on barium meal indicating mucosal oedema associated with ulcer LIVER Councilman Body eosinophilic globules in liver An inclusion found in the cytoplasm of liver cells. Mallory bodies are damaged intermediate Mallory Body filaments within the hepatocytes BILIARY Boas' Sign Dermal hyperaesthesia at inferior angle of R scapula Courvoisier's Law Palpable gall bladder w/ painless jaundice unlikely to be cholelithiasis ACCESSORY Charcot's Jaundice, fever and chills, RUQ pain Cholangitis -

Diagnostic Role of Procalcitonin in Neonatal Sepsis In
DIAGNOSTIC ROLE OF PROCALCITONIN IN NEONATAL SEPSIS IN TERTIARY CARE HOSPITAL DISSERTATION SUBMITTED TO In partial fulfillment of the requirement for the degree of DOCTOR OF MEDICINE IN MICROBIOLOGY (Branch IV) M. D. (MICROBIOLOGY) of THE TAMIL NADU DR. M. G. R MEDICAL UNIVERSITY CHENNAI- 600032 DEPARTMENT OF MICROBIOLOGY TIRUNELVELI MEDICAL COLLEGE TIRUNELVELI- 11 MAY 2018 BONAFIDE CERTIFICATE This is to certify that the dissertation entitled “Diagnostic role of Procalcitonin in neonatal sepsis in tertiary care hospital” submitted by Dr.D.Jeyaganguli to the Tamilnadu Dr. M.G.R Medical University, Chennai, in partial fulfillment of the requirement for the award of M.D. Degree Branch – IV (Microbiology) is a bonafide research work carried out by her under direct supervision & guidance. Head of the Department, Department of Microbiology Tirunelveli Medical College, Tirunelveli. CERTIFICATE This is to certify that the Dissertation “Diagnostic role of procalcitonin in neonatal sepsis in tertiary care hospital” presented here in by Dr.D.Jeyaganguli is an original work done in the Department of Microbiology, Tirunelveli Medical College Hospital,Tirunelveli for the award of Degree of M.D. (Branch IV) Microbiology under my guidance and supervision during the academic period of 2015 -2018. The DEAN Tirunelveli Medical College, Tirunelveli - 627011. DECLARATION I solemnly declare that the dissertation titled“Diagnostic role of procalcitonin in neonatal sepsis in tertiary care hospital” is done by me at Tirunelveli Medical College hospital, Tirunelveli. I also declare that this bonafide work or a part of this work was not submitted by me or any others for any award, degree, or diploma to any other University, Board, either in or abroad.