US 2012/0244131 A1 Delacote Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Investigation of Candidate Genes and Mechanisms Underlying Obesity
Prashanth et al. BMC Endocrine Disorders (2021) 21:80 https://doi.org/10.1186/s12902-021-00718-5 RESEARCH ARTICLE Open Access Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules G. Prashanth1 , Basavaraj Vastrad2 , Anandkumar Tengli3 , Chanabasayya Vastrad4* and Iranna Kotturshetti5 Abstract Background: Obesity associated type 2 diabetes mellitus is a metabolic disorder ; however, the etiology of obesity associated type 2 diabetes mellitus remains largely unknown. There is an urgent need to further broaden the understanding of the molecular mechanism associated in obesity associated type 2 diabetes mellitus. Methods: To screen the differentially expressed genes (DEGs) that might play essential roles in obesity associated type 2 diabetes mellitus, the publicly available expression profiling by high throughput sequencing data (GSE143319) was downloaded and screened for DEGs. Then, Gene Ontology (GO) and REACTOME pathway enrichment analysis were performed. The protein - protein interaction network, miRNA - target genes regulatory network and TF-target gene regulatory network were constructed and analyzed for identification of hub and target genes. The hub genes were validated by receiver operating characteristic (ROC) curve analysis and RT- PCR analysis. Finally, a molecular docking study was performed on over expressed proteins to predict the target small drug molecules. Results: A total of 820 DEGs were identified between -

Clinical and Immunological Diversity of Recombination Defects Hanna
Clinical and Immunological Diversity Defects of Recombination Hanna IJspeert Clinical and immunological diversity of recombination defects Hanna IJspeert 2014 Clinical and Immunological Diversity of Recombination Defects Hanna IJspeert The research for this thesis was performed within the framework of the Erasmus Postgraduate School Molecular Medicine. The studies described in the thesis were performed at the Department of Immunology, Erasmus MC, University Medical Center Rotterdam, Rotterdam, the Netherlands and collaborating institutions. The studies were financially suported by ‘Sophia Kinderziekhuis Fonds’ (grant 589). The printing of this thesis was supported by Erasmus MC, Stichting Kind & Afweer, CSL Behring and BD Biosciences. ISBN: 978-94-91811-04-3 Illustrations: Sandra de Bruin-Versteeg, Hanna IJspeert Cover: Stephanie van Brandwijk Lay-out: Caroline Linker Printing: Haveka B.V., Alblasserdam, the Netherlands Copyright © 2014 by Hanna IJspeert. All rights reserved. No part of this book may be reproduced, stored in a retrieval system of transmitted in any form or by any means, without prior permission of the author. Clinical and Immunological Diversity of Recombination Defects Klinische en immunologische diversiteit van recombinatie defecten Proefschrift ter verkrijging van de graad van doctor aan de Erasmus Universiteit Rotterdam op gezag van de rector magnificus Prof.dr. H.A.P. Pols en volgens besluit van het College voor Promoties. De openbare verdediging zal plaatsvinden op woensdag 19 maart 2014 om 13.30 uur door Hanna IJspeert geboren te Dordrecht PROMOTIE COMMISSIE Promotoren Prof.dr. J.J.M. van Dongen Prof.dr. A.J. van der Heijden Overige leden Prof.dr. B.H. Gaspar Prof.dr. F.J.T. Staal Dr. -

United States Patent (10) Patent No.: US 9,044,492 B2 Delacote Et Al
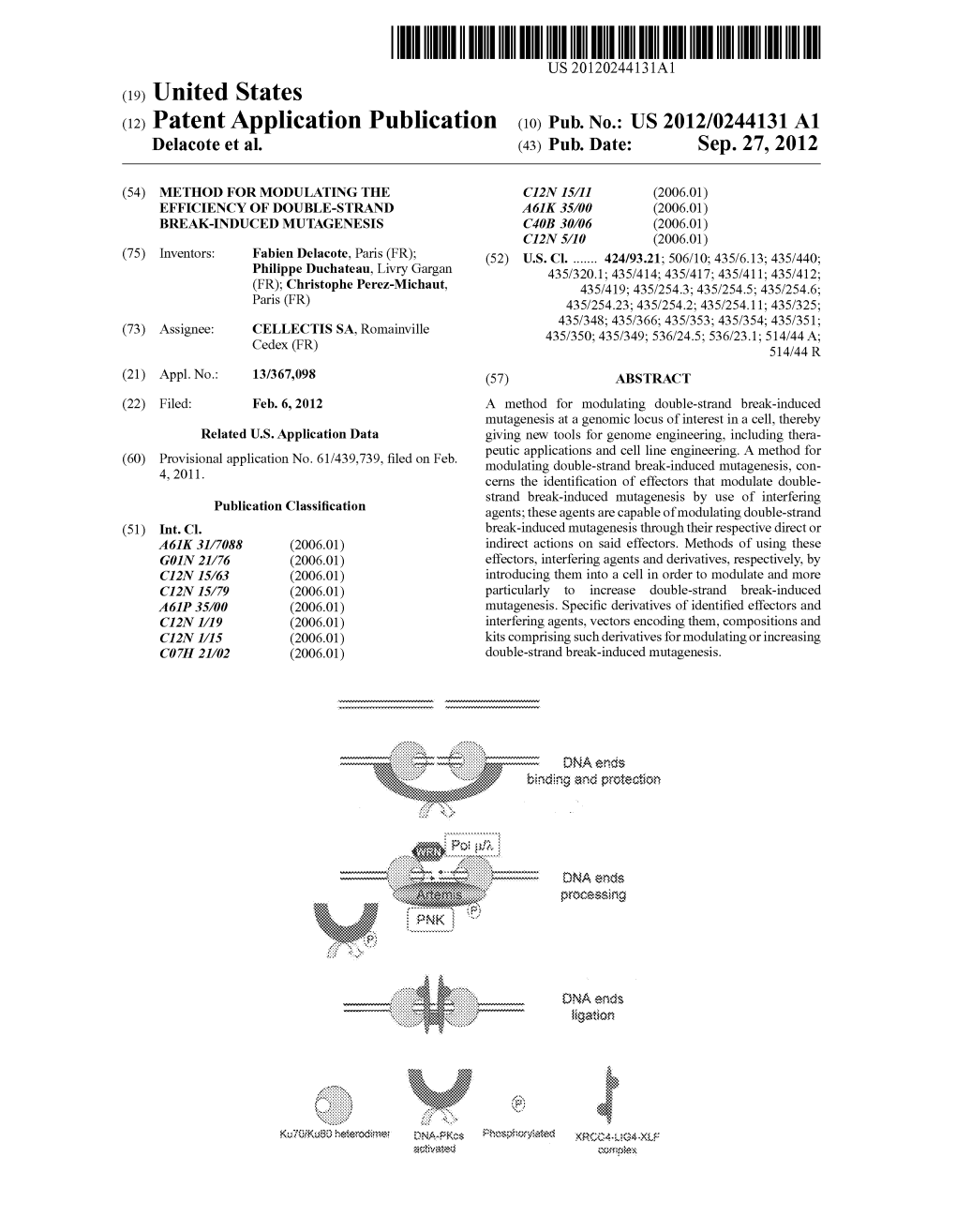
USOO9044492B2 (12) United States Patent (10) Patent No.: US 9,044,492 B2 Delacote et al. (45) Date of Patent: Jun. 2, 2015 (54) METHOD FORMODULATING THE 8,206,965 B2 6/2012 Arnould et al. EFFICIENCY OF DOUBLE-STRAND 8,211,685 B2 7/2012 Epinatet al. 8.426,177 B2 4/2013 Gouble BREAK-INDUCED MUTAGENESIS 8,476,072 B2 7/2013 Cabaniols et al. O O 8,530,214 B2 9/2013 Arnould et al. (75) Inventors: Fabien Delacote, Paris (FR); Philippe 2006, OO78552 A1 4/2006 Arnould et al. Duchateau, Livry Gargan (FR): 2006/0153826 A1 7/2006 Arnould et al. Christophe Perez-Michaut, Paris (FR) 2006/0206949 A1 9, 2006 Arnould et al. s 2009/0220476 A1 9/2009 Paques 2009/0222937 A1 9, 2009 Arnould et al. (73) Assignee. CELLECTISSA, Romainville (FR) 2009/0271881 A1 10, 2009 Arnould et al. - 2010/0086533 A1 4/2010 Montoya et al. (*) Notice: Subject to any disclaimer, the term of this 2010, 0146651 A1 6, 2010 Smith et al. patent is extended or adjusted under 35 2010, 0151556 Al 6, 2010 Arnould et al. U.S.C. 154(b) by 407 days. 2010.0167357 A1 7/2010 Fajardo Sanchez et al. 2010/02O3031 A1 8, 2010 Grizot et al. (21) Appl. No.: 13/367,098 2010/0229252 A1 9/2010 Perez-Michaut 9 (Continued) (22) Filed: Feb. 6, 2012 Primary Examiner — Terra C Gibbs (65) Prior Publication Data (74) Attorney, Agent, or Firm — Oblon, McClelland, US 2012/0244131 A1 Sep. 27, 2012 Maier & Neustadt, L.L.P. Related U.S. -

Polymerase Is a Robust Terminal Transferase That Oscillates Between
RESEARCH ARTICLE Polymerase is a robust terminal transferase that oscillates between three different mechanisms during end-joining Tatiana Kent1,2, Pedro A Mateos-Gomez3,4, Agnel Sfeir3,4, Richard T Pomerantz1,2* 1Fels Institute for Cancer Research, Temple University Lewis Katz School of Medicine, Philadelphia, United States; 2Department of Medical Genetics and Molecular Biochemistry, Temple University Lewis Katz School of Medicine, Philadelphia, United States; 3Skirball Institute of Biomolecular Medicine, New York University School of Medicine, New York, United States; 4Department of Cell Biology, New York University School of Medicine, New York, United States Abstract DNA polymerase q (Polq) promotes insertion mutations during alternative end-joining (alt-EJ) by an unknown mechanism. Here, we discover that mammalian Polq transfers nucleotides to the 3’ terminus of DNA during alt-EJ in vitro and in vivo by oscillating between three different modes of terminal transferase activity: non-templated extension, templated extension in cis, and templated extension in trans. This switching mechanism requires manganese as a co-factor for Polq template-independent activity and allows for random combinations of templated and non- templated nucleotide insertions. We further find that Polq terminal transferase activity is most efficient on DNA containing 3’ overhangs, is facilitated by an insertion loop and conserved residues that hold the 3’ primer terminus, and is surprisingly more proficient than terminal deoxynucleotidyl transferase. In summary, this report identifies an unprecedented switching mechanism used by Polq to generate genetic diversity during alt-EJ and characterizes Polq as among the most proficient terminal transferases known. DOI: 10.7554/eLife.13740.001 *For correspondence: richard. -

Mechanism of Genome Instability Mediated by Human DNA Polymerase Mu Misincorporation
ARTICLE https://doi.org/10.1038/s41467-021-24096-7 OPEN Mechanism of genome instability mediated by human DNA polymerase mu misincorporation Miao Guo1,2, Yina Wang1,2, Yuyue Tang1,2, Zijing Chen1,2, Jinfeng Hou1,2, Jingli Dai 1,2, Yudong Wang1,2, ✉ ✉ Liangyan Wang1,2, Hong Xu1,2, Bing Tian1,2, Yuejin Hua 1,2 & Ye Zhao 1,2 Pol μ is capable of performing gap-filling repair synthesis in the nonhomologous end joining (NHEJ) pathway. Together with DNA ligase, misincorporation of dGTP opposite the tem- 1234567890():,; plating T by Pol μ results in a promutagenic T:G mispair, leading to genomic instability. Here, crystal structures and kinetics of Pol μ substituting dGTP for dATP on gapped DNA sub- strates containing templating T were determined and compared. Pol μ is highly mutagenic on a 2-nt gapped DNA substrate, with T:dGTP base pairing at the 3ʹ end of the gap. Two residues (Lys438 and Gln441) interact with T:dGTP and fine tune the active site micro- environments. The in-crystal misincorporation reaction of Pol μ revealed an unexpected second dGTP in the active site, suggesting its potential mutagenic role among human X family polymerases in NHEJ. 1 Institute of Biophysics, College of Life Sciences, Zhejiang University, Hangzhou, Zhejiang, China. 2 MOE Key Laboratory of Biosystems Homeostasis & ✉ Protection, Zhejiang University, Hangzhou, Zhejiang, China. email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3759 | https://doi.org/10.1038/s41467-021-24096-7 | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-24096-7 ccurate DNA replication upon DNA damage by DNA templating base, which may easily cause microhomology-directed Apolymerases is a key factor determining genome stability deletions19. -

Supplemental Data
Article TCF7L2 is a master regulator of insulin production and processing ZHOU, Yuedan, et al. Abstract Genome-wide association studies have revealed >60 loci associated with type 2 diabetes (T2D), but the underlying causal variants and functional mechanisms remain largely elusive. Although variants in TCF7L2 confer the strongest risk of T2D among common variants by presumed effects on islet function, the molecular mechanisms are not yet well understood. Using RNA-sequencing, we have identified a TCF7L2-regulated transcriptional network responsible for its effect on insulin secretion in rodent and human pancreatic islets. ISL1 is a primary target of TCF7L2 and regulates proinsulin production and processing via MAFA, PDX1, NKX6.1, PCSK1, PCSK2 and SLC30A8, thereby providing evidence for a coordinated regulation of insulin production and processing. The risk T-allele of rs7903146 was associated with increased TCF7L2 expression, and decreased insulin content and secretion. Using gene expression profiles of 66 human pancreatic islets donors', we also show that the identified TCF7L2-ISL1 transcriptional network is regulated in a genotype-dependent manner. Taken together, these results demonstrate that not only synthesis of [...] Reference ZHOU, Yuedan, et al. TCF7L2 is a master regulator of insulin production and processing. Human Molecular Genetics, 2014, vol. 23, no. 24, p. 6419-6431 DOI : 10.1093/hmg/ddu359 PMID : 25015099 Available at: http://archive-ouverte.unige.ch/unige:45177 Disclaimer: layout of this document may differ from the published -

Development and Characterization of Carboplatin, Docetaxel, and Combined Carboplatin/Docetaxel Resistant Ovarian Cancer Cell Lines
DEVELOPMENT AND CHARACTERIZATION OF CARBOPLATIN, DOCETAXEL, AND COMBINED CARBOPLATIN/DOCETAXEL RESISTANT OVARIAN CANCER CELL LINES by STEPHEN ROBERT ARMSTRONG Thesis submitted as a partial requirement in the Master of Science (M.Sc.) in Biology School of Graduate Studies Laurentian University Sudbury, Ontario © Stephen R. Armstrong, 2010 Library and Archives Bibliothgque et 1*1 Canada Archives Canada Published Heritage Direction du Branch Patrimoine de l'6dition 395 Wellington Street 395, rue Wellington Ottawa ON K1A0N4 Ottawa ON K1A 0N4 Canada Canada Your file Votra r6f6rence ISBN: 978-0-494-70639-8 Our file Notre r6f6rence ISBN: 978-0-494-70639-8 NOTICE: AVIS: The author has granted a non- L'auteur a accorde une licence non exclusive exclusive license allowing Library and permettant a la Bibliotheque et Archives Archives Canada to reproduce, Canada de reproduce, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par I'lnternet, preter, telecommunication or on the Internet, distribuer et vendre des theses partout dans le loan, distribute and sell theses monde, a des fins commerciales ou autres, sur worldwide, for commercial or non- support microforme, papier, electronique et/ou commercial purposes, in microform, autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in this et des droits moraux qui protege cette these. Ni thesis. Neither the thesis nor la these ni des extraits substantiels de celle-ci substantial extracts from it may be ne doivent etre imprimes ou autrement printed or otherwise reproduced reproduits sans son autorisation. -

25% OFF Full-Size Primary Antibody
GeneTex Antibodies with Citations - 25% OFF full-size Primary Antibody List of Eligible Products Cat No. Product Name GTX130274 CD31 antibody GTX127309 HIF1 alpha antibody GTX635679 SARS-CoV-2 (COVID-19) nucleocapsid antibody [HL344] GTX632604 SARS-CoV / SARS-CoV-2 (COVID-19) spike antibody [1A9] GTX42110 CD3 epsilon antibody [CD3-12] GTX31878 SLC27A6 antibody GTX101125 LPL antibody GTX100118 GAPDH antibody GTX118000 STAT3 (phospho Tyr705) antibody GTX53048 ULBP2 antibody [7F33] GTX130422 IRF3 (phospho Ser386) antibody GTX121677 Histone H3K9me3 (Tri-methyl Lys9) antibody GTX101452 DYNC1H1 antibody GTX86952 Caspase 3 (cleaved Asp175) antibody GTX85378 CUEDC1 antibody GTX84785 BTK antibody [10E10] GTX84377 HDAC6 antibody [4C5] GTX77457 DDDDK tag antibody GTX76185 CD9 antibody [MM2/57] GTX76060 CD11b antibody [OX-42] GTX74034 IL1 beta antibody GTX70485 Ku80 antibody GTX70268 Hec1 antibody [9G3.23] GTX632426 Iba1 antibody [GT10312] GTX629630 beta Actin antibody [GT5512] GTX629117 Dengue virus Envelope protein antibody [GT643] GTX628802 alpha Tubulin antibody [GT114] GTX627408 GAPDH antibody [GT239] GTX60661 ABCB5 antibody [5H3C6] GTX59618 ERK1/2 antibody GTX55142 COL11A1 antibody GTX54648 Cruciform DNA antibody [2D3] GTX54088 HMGCR antibody GTX54007 FBP1 antibody GTX51959 SRX1 antibody GTX50128 AKT (phospho Ser473) antibody GTX50090 GSK3 beta (phospho Ser9) antibody GTX41877 CD63 antibody [AD1] GTX41794 CD81 antibody [Eat2] GTX41286 Collagen I antibody GTX39744 Marco antibody [ED31] GTX32224 IKB alpha (phospho Ser32/Ser36) antibody GTX31308 -

Essential Role for Polymerase Specialization in Cellular PNAS PLUS Nonhomologous End Joining
Essential role for polymerase specialization in cellular PNAS PLUS nonhomologous end joining John M. Pryora,1, Crystal A. Watersa,1, Ana Azab, Kenjiro Asagoshia, Christina Stroma, Piotr A. Mieczkowskic, Luis Blancob, and Dale A. Ramsdena,2 aDepartment of Biochemistry and Biophysics and Curriculum in Genetics and Molecular Biology, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC 27599; bCentro de Biología Molecular Severo Ochoa (Consejo Superior de Investigaciones Científicas-Universidad Autonoma de Madrid), 28049 Madrid, Spain; and cDepartment of Genetics, University of North Carolina, Chapel Hill, NC 27599 Edited by Graham C. Walker, Massachusetts Institute of Technology, Cambridge, MA, and approved July 1, 2015 (received for review March 23, 2015) Nonhomologous end joining (NHEJ) repairs chromosome breaks (Loop1) (9, 11, 12). However, evidence that these different in vitro and must remain effective in the face of extensive diversity in activities effects the biological role of these polymerases is definitive broken end structures. We show here that this flexibility is often only for TdT, which adds template-independent nucleotides reliant on the ability to direct DNA synthesis across strand breaks, (N additions) to broken intermediates during V(D)J recombination. and that polymerase (Pol) μ and Pol λ are the only mammalian Accordingly, TdT is expressed only in cells active in V(D)J re- DNA polymerases that have this activity. By systematically varying combination (13). substrate in cells, we show each polymerase is uniquely proficient By comparison, both Pol μ and Pol λ are widely expressed (14, in different contexts. The templating nucleotide is also selected 15), and it has been difficult to clearly link phenotypes of de- differently, with Pol μ using the unpaired base adjacent to the ficiency in these polymerases to an important role for them in downstream 5′ phosphate even when there are available template NHEJ. -

Small‐Molecule Inhibitors of Dna Polymerase Function
SMALL‐MOLECULE INHIBITORS OF DNA POLYMERASE FUNCTION DISSERTATION zur Erlangung des Akademischen Grades des Doktors der Naturwissenschaften (Dr. rer. nat.) vorgelegt von Tobias Strittmatter aus Küssaberg‐Kadelburg an der Universität Konstanz Mathematisch‐Naturwissenschaftliche Sektion Fachbereich Chemie 2014 Tag der mündlichen Prüfung: 05. Dezember 2014 Prüfungsvorsitz: Prof. Dr. Gerhard Müller 1. Referent: Prof. Dr. Andreas Marx 2. Referent: Prof. Dr. Thomas U. Mayer Konstanzer Online-Publikations-System (KOPS) URL: http://nbn-resolving.de/urn:nbn:de:bsz:352-0-274434 „Meinen Eltern” Danksagung Die vorliegende Arbeit entstand in der Zeit von November 2009 bis August 2014 in der Arbeitsgruppe von Prof. Dr. Andreas Marx am Lehrstuhl für Organische und Zelluläre Chemie am Fachbereich Chemie der Universität Konstanz. Nach diesem Zeitraum geprägt von intensiver Forschungsarbeit liegt nun meine Dissertation vor und ein weiteres Kapitel meiner beruflichen Karriere kann nun erfolgreich abgeschlossen werden. Aus diesem Grund ist es jetzt auch an der Zeit, mich bei all denen zu bedanken, die mich während dieser spannenden Zeit begleitet, unterstützt und gefördert haben. Meinem Doktorvater Prof. Dr. Andreas Marx danke ich ganz herzlich für die Überlassung der sehr interessanten und interdisziplinären Themenstellung, sowie für das in mich gesetzte Vertrauen, welches mir viel Raum für die selbstständige Bearbeitung und kreative Gestaltung des Themas erlaubte. Ihm, wie auch den weiteren Mitgliedern meines „Thesis Committee“, Prof. Dr. Thomas Mayer und Herrn Prof. Dr. Michael Berthold, danke ich zudem für die anregenden wissenschaftlichen Diskussionen und die geleisteten Hilfestellungen. Prof. Dr. Thomas Mayer danke ich außerdem für die Übernahme des Zweitgutachtens und Herrn Prof. Dr. Gerhard Müller für die Übernahme des Prüfungsvorsitzes. -

DNA Methylome Distinguishes Oropharyngeal and Oral Cancer from Oral Lesions and Healthy Oral Mucosa
DNA Methylome Distinguishes Oropharyngeal and Oral Cancer from Oral Lesions and Healthy Oral Mucosa Milutin Gašperov Nina ( [email protected] ) Institut Ruder Boskovic Ivan Sabol Institut Ruder Boskovic Ksenija Božinović Institut Ruder Boskovic Emil Dediol Klinicka Bolnica Dubrava Marinka Mravak-Stipetić Sveuciliste u Zagrebu Stomatoloski fakultet Danilo Licastro CBM Scrl Simeone Dal Monego CBM Scrl Magdalena Grce ( [email protected] ) Institut Ruder Boskovic https://orcid.org/0000-0001-6178-8418 Research Keywords: DNA methylation, head and neck squamous cell carcinoma (HNSCC), potentially premalignant oral lesions, healthy oral mucosa, human papillomavirus (HPV) Posted Date: April 3rd, 2020 DOI: https://doi.org/10.21203/rs.3.rs-17778/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/30 Abstract Background: There is a strong need to nd new, good biomarkers of head and neck squamous cell carcinoma (HNSCC), because of the prognoses and high mortality of patients. The aim of this study was to identify the potential biomarkers in HNSCC that have differences in their DNA methylome and potentially premalignant oral lesions, in comparison to healthy oral mucosa. Methods: In this study 32 oral samples were tested: 9 healthy oral mucosae, 13 HNSCC and 10 potentially premalignant oral lesions for DNA methylation by Innium MethylationEPIC BeadChip. Results: Our ndings showed a panel of genes signicantly hypermethylated in their promoters or specic sites in HNSCC samples in comparison to healthy oral samples, which are mainly oncogenes, receptor and transcription factor genes, or genes included in cell cycle, transformation, apoptosis and autophagy. A group of hypomethylated genes in HNSCC, in comparison to healthy oral mucosa, is mainly involved in the host immune response and transcriptional regulation. -

DNA Polymerases and Human Disease
REVIEWS Nature Reviews Genetics | AOP, published online 15 July 2008; doi:10.1038/nrg2345 DNA polymerases and human disease Lawrence A. Loeb*‡§ and Raymond J. Monnat Jr*|| Abstract | The human genome encodes at least 14 DNA-dependent DNA polymerases — a surprisingly large number. These include the more abundant, high-fidelity enzymes that replicate the bulk of genomic DNA, together with eight or more specialized DNA polymerases that have been discovered in the past decade. Although the roles of the newly recognized polymerases are still being defined, one of their crucial functions is to allow synthesis past DNA damage that blocks replication-fork progression. We explore the reasons that might justify the need for so many DNA polymerases, describe their function and mode of regulation, and finally consider links between mutations in DNA polymerases and human disease. non-processive Lagging strand Our knowledge of human DNA polymerases has under- low catalytic efficiency and are . Eight to One of the two DNA strands gone a striking expansion in the past decade. Since ten of these TLS DNA polymerases seem to be present that is synthesized during DNA the discovery in 1957 of an enzyme that catalyses the in most human cells (FIG. 1b; TABLE 1) and are probably replication. The lagging strand accurate replication of DNA, there has been a progres- found in all mammalian cells. Three TLS polymerases is synthesized by Pol δ in short segments that are known as sive accumulation of evidence for five ‘classical’ DNA have been identified in Saccharomyces cerevisiae, and 14 Okasaki fragments. polymerases in all mammalian cells, each functioning two are known in Escherichia coli .