A Quick and Sensitive Diagnostic Tool for Detection of Maize Streak Virus Mathias Tembo1*, Adedapo O
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
REPORT of the AUDITOR – GENERAL on the ACCOUNTS
REPORT of the AUDITOR – GENERAL ON THE ACCOUNTS FOR THE FINANCIAL YEAR ENDED 31st DECEMBER 2007 2 TABLE OF CONTENTS Page Introduction..................................................................................................... 1 Audit Scope and Methodology....................................................................... 1 Institutional Development.............................................................................. 1 International Co-operation............................................................................ 1 Accountability of Public Funds...................................................................... 2 Limitation of Scope....................................................................................... 2 Outturn and Appropriation Accounts............................................................ 2 General Revenues.......................................................................................... 3 Zambia Revenue Authority........................................................................... 3 Exceptional Revenue – Ministry of Energy and Water Development........... 6 Fees and Fines – Ministry of Homes Affairs – Police ................................. 7 Exceptional Revenue – Ministry of Agriculture and Cooperatives.............. 9 Fees and Fines - Ministry of Energy and Water – Water Board.................. 9 Fees and Fines – Ministry of Mines and Mineral Development.................. 10 Fees and Fines – Ministry of Home Affairs – Immigration....................... 12 Fees and -

05 February 2021
ZAMBIA COVID-19 SITUATION REPORT NO. 132 th th Disease Pandemic: COVID-19 Response start date: 30 January, 2020 Outbreak Declared:18 March, 2020 Report date: Friday 5th February 2021 Prepared by: MOH/ZNPHI/WHO Correspondence:[email protected] 1. SITUATION UPDATE This week (1st - 7th Feb) Cases 6,210 Deaths 65 Recoveries 4,045 1.1 CURRENT CASE NUMBERS (as of 09:00 hours CAT) • In the past 24 hrs, we recorded 1,424 new confirmed cases, 16 deaths and Global Numbers 740 recoveries. (Source: JHU) 104,886,168 Confirmed • Cumulative number of confirmed COVID-19 cases recorded to date is 60,427 2,284,686 Deaths (2.2% CFR) 58,322,664 Recoveries with 828 deaths (CFR=1.37%) and 52,045 recoveries (86.13% recovered). Africa Numbers • Of the 828 total deaths among the confirmed cases, 378 have been classified Source: Africa CDC) 3,626,960 Confirmed as COVID-19 deaths (CFR=0.63%) and 419 as associated deaths; 31 deaths 93,647 Deaths (2.6% CFR) are pending classification. See Annex 1 for definitions 3,128,534 Recoveries • There are currently 7,554 active cases: of these, 409 (5.4%) are hospitalised (with 285 on Oxygen therapy and 37 in critical condition); 7,145 patients are under community management. 2. EPIDEMIOLOGICAL HIGHLIGHTS 14000 12000 10000 8000 6000 Number Recorded Number 4000 2000 0 5-11Oct 1 -1Feb7 7-13Sep 3 -Aug39 2 -2Nov8 4 -410 Jan 19-25 Oct 12-18 Oct 13-19 Apr 20-26 Apr 21-27 Sep 9 -915Nov 15 15 -21Jun 6 -612 Apr 16-22 Mar 23-29 Mar 14 14 Sep-20 -713 Dec 1 - 17 Jun 13 13 July-19 8 -814 Jun 11 11 - 17 Jan 25 - 31 Jan 18 18 - 24 Jan -

USAID/Zambia Partners in Development Book
PARTNERS IN DEVELOPMENT July 2018 Partners in Zambia’s Development Handbook July 2018 United States Agency for International Development Embassy of the United States of America Subdivision 694 / Stand 100 Ibex Hill Road P.O. Box 320373 Lusaka, Zambia 10101 Cover Photo: As part of a private -sector and youth-engagement outreach partnership, media entrepreneur and UNAIDS Ambassador Lulu Haangala Wood (l), musician and entrepreneur Pompi (c), and Film and TV producer Yoweli Chungu (r) lend their voices to help draw attention to USAID development programs. (Photo Credit: Chando Mapoma / USAID Zambia) Our Mission On behalf of the American People, we promote and demonstrate democratic values abroad, and advance a free, peaceful, and prosperous world. In support of America's foreign policy, the U.S. Agency for International Development leads the U.S. Government's international development and disaster assistance through partnerships and investments that save lives, reduce poverty, strengthen democratic governance, and help people emerge from humanitarian crises and progress beyond assistance. Our Interagency Initiatives USAID/Zambia Partners In Development 1 The United States Agency for International Development (USAID) is the lead U.S. Government agency that works to end extreme global poverty and enable resilient, democratic societies to realize their potential. Currently active in over 100 countries worldwide, USAID was born out of a spirit of progress and innovation, reflecting American values and character, motivated by a fundamental belief in helping others. USAID provides development and humanitarian assistance in Africa, Asia and the Near East, Latin America and the Caribbean, and Europe. Headquartered in Washington, D.C., USAID retains field missions around the world. -

Familiarisation Tour of Mpulungu, Zambia
THE ENVIRONMENTAL COUNCIL OF ZAMBIA Pollution Control and Other Measures to protect Biodiversity in Lake Tanganyika (RAF/92/G32) FAMILIARISATION TOUR OF MPULUNGU A COMBINED SOCIO-ECONO0MICS AND ENVIRONMNETAL EDUCATION TOUR CONDUCTED FROM 2/2/99 TO 3/3/99 Munshimbwe Chitalu Assistant National Co-ordinator Socio-economics Co-ordinator National Coordination Office LUSAKA ZAMBIA July 2000 M p u l u n g u Vi s i t R e p o r t , So c i o - E c o n o m i c s / E n v i r o n m e n t a l Ed u c a t i o n Contents List of Acronyms ii Foreword iii Executive summary iv 1 HIGHLIGHTS 1 1 Environmental Education Activities 1 2 Conservation and Development Committees 1 3 Activities of CDCs 2 4 National Project coordination 3 5 The team 3 6 Approach and salutations 3 2 THE TOUR IN MORE DETAIL 4 1 The Aim 4 2 Specific Objectives 4 3 Findings 4 3.1 Community Development Officer (CDO) 4 3.2 Department of Fisheries (DoF) 5 3.3 Immigration Department 7 3.4 Mpulungu District Council 7 3.5 Mpulungu Harbor Corporation Limited 8 3.6 Mr. Mugala 8 3.7 The Provincial Agricultural Co-ordination Office (PACO) 9 3.8 Police Service 9 3.9 Senior Chief Tafuna 9 3.10 Stratum 2 CDC 9 3.11 Village CDCs 10 3 CONCLUSIONS AND RECOMMENDATIONS 12 4 PROPOSED IMMEDIATE ACTIONS 14 Appendix I: Institutions and individuals visited 15 Appendix II: Itinerary 17 Appendix III: Resources 18 P A G E I M p u l u n g u Vi s i t R e p o r t , So c i o - E c o n o m i c s / E n v i r o n m e n t a l Ed u c a t i o n List of Acronyms AMIS Association of Micro-finance Institutions of Zambia ANSEC -

TEVETA, RTSA Launch Instructor Driving Curriculum
News Issue No. 1 Volume January 11 No. 2 - MarchApr 2017 - Jun 2013 TEVETA, RTSA Launch Instructor Driving Government to turn ZAMIM City Campus into Curriculum Business Incubator Centre for TEVET Graduates • Prisoners find hope in TEVET: The case of skills training at Mukobeko Maximum INSIDE •• 2017“The –2019Disease TEVETA is still here,Strategic let’s fightPlan on” • BEAR Project ends: A look at its milestones THISTEVETTEVET News Newsletter 2012 • From Kabwe Trades to Kabwe Institute of Technology: What does it entail? 1 1 ISSUE • Curriculum review: A component of the SSTEP project TEVET 1st Quarter Newsletter 2017 | 1 CONTENTS FORWORD Foreword 2 e welcome you to the 2017 fisrt Quarter TEVET TEVETA, RTSA Launch Instructor Driving 3 newsletter. 2017 started Curriculum W with valuable landmarks among 2017 –2019 TEVETA strategic 2017 –2019 TEVETA Strategic Plan: TEVET for 4 plan. The Strategic Plan is aligned Internationally competitive and transversally to the Republican President’s skilled persons message during the opening of the First Session of the 12th National Assembly that “my administration BEAR project ends: A look at its milestones 5 will create an atmosphere where sectors will work together simultaneously to resolve developmental Northern Technical College graduate wins challenges such as youth unemployment and high levels of poverty, by 7 innovation award harnessing the youthful population into a productive one. This entails the youth embracing innovation and entrepreneurship, advanced technologies and actively participating in the economy.” Curriculum review: A component of the SSTEP 7 project During the quarter, TEVETA and the Road, Transport and Safety Agency (RTSA) launched a driving curriculum for driving instructors. -
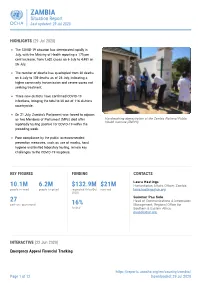
ZAMBIA Situation Report Last Updated: 29 Jul 2020
ZAMBIA Situation Report Last updated: 29 Jul 2020 HIGHLIGHTS (29 Jul 2020) The COVID-19 situation has deteriorated rapidly in July, with the Ministry of Health reporting a 175 per cent increase, from 1,632 cases on 6 July to 4,481 on 26 July. The number of deaths has quadrupled from 30 deaths on 6 July to 139 deaths as of 26 July, indicating a higher community transmission and severe cases not seeking treatment. Three new districts have confirmed COVID-19 infections, bringing the total to 38 out of 116 districts countrywide. On 21 July, Zambia’s Parliament was forced to adjourn as two Members of Parliament (MPs) died after Handwashing demostration at the Zambia National Public Health Institute (ZNPHI) reportedly testing positive for COVID-19 within the preceding week. Poor compliance by the public to recommended prevention measures, such as use of masks, hand hygiene and limited laboratory testing, remain key challenges to the COVID-19 response. KEY FIGURES FUNDING CONTACTS Laura Hastings 10.1M 6.2M $132.9M $21M Humanitarian Affairs Officer, Zambia people in need people targeted requested (May-Oct received [email protected] 2020) Guiomar Pau Sole 27 Head of Communications & Information partners operational 16% Management, Regional Office for funded Southern & Eastern Africa [email protected] INTERACTIVE (22 Jun 2020) Emergency Appeal Financial Tracking https://reports.unocha.org/en/country/zambia/ Page 1 of 12 Downloaded: 29 Jul 2020 ZAMBIA Situation Report Last updated: 29 Jul 2020 View this interactive graphic: https://bit.ly/ZambiaAppeal2020Funding BACKGROUND (29 Jul 2020) Situation Overview Zambia recorded the first case of COVID-19 on 18 March 2020, and as 26 July 4,481 cases had been confirmed with 139 deaths reported, in 38 out of 116 districts countrywide, according to the Ministry of Health (MoH). -
C:\Users\Public\Documents\GP JOBS\Gazette No. 73 of Friday, 16Th
REPUBLIC OF ZAMBIA Price: K5 net Annual Subscription: Within Lusaka—K200.00 Published by Authority Outside Lusaka—K230.00 No. 6430] Lusaka, Friday, 16th October, 2015 [Vol. LI, No. 73 GAZETTE NOTICE NO. 643 OF 2015 [5929855/13 Zambia Information and Communications Technologies Authority The Information and Communications Technologies Act, 2009 (Act No. 15 of 2009) Notice of Determination of Unserved and Underserved Areas Section 70 (2) of the Information and Communication TechnologiesAct No. 15 of 2009 (ICTAct) empowers the Zambia Information and Communications Technology Authority (ZICTA) to determine a system to promote the widespread availability and usage of electronic communications networks and services throughout Zambia by encouraging the installation of electronic communications networks and the provision for electronic communications services in unserved and underserved areas and communities. Further, Regulation 5 (2) of Statutory Instrument No. 38 of 2012 the Information and Communications Technologies (Universal Access) Regulations 2012 mandates the Authority to designate areas as universal service areas by notice in the gazette. In accordance with the said regulations, the Authority hereby notifies members of the public that areas contained in the Schedule Hereto are hereby designated as universal service areas. M. K. C. MUDENDA (MRS.) Director General SN Site Name Longtitude Latitude Elevation Province 1 Nalusanga_Chunga Headquarter Offices 27.22415 -15.22135 1162 Central 2 Mpusu_KankamoHill 27.03507 -14.45675 1206 Central -

Kenya - Caucus Parliamentary Parliamentary National Assembly of Kenya National Assembly Hon
AFRICA 2019-2020 THE ICCF GROUP INTERNATIONALCONSERVATION.ORG COPYRIGHT INTERNATIONAL CONSERVATION CAUCUS FOUNDATION MISSION THE MOST ADVANCED TO ADVANCE SOLUTION IN CONSERVATION CONSERVATION GOVERNANCE BY WE BUILD POLITICAL WILL BUILDING The ICCF Group advances governments' leadership in conservation internationally by building political will POLITICAL WILL, within legislatures and supporting governments in the management of protected areas. PROVIDING ON-THE-GROUND CATALYZING CHANGE WITH KNOWLEDGE & EXPERTISE SOLUTIONS The ICCF Group supports political will by catalyzing strategic partnerships and knowledge sharing between policymakers and our extensive network. VISION TO PRESERVE THE WORLD'S MOST CRITICAL LANDSCAPES A WORLD The ICCF Group's international track record in legislative outcomes, public-private partnerships, & land WHERE management demonstrates that our unique model is emerging as a cost-effective, sustainable solution to PEOPLE AND conservation governance challenges. NATURE SUSTAIN AND NURTURE ONE ANOTHER 2019-2020 THE ICCF GROUP THE ICCF GROUP IN AFRICA EMPOWERING POLICYMAKERS &AND THE JUDICIARY The ICCF Group is working in East, Southern, and Central Africa to foster political will for conservation and support the sustainable management of natural resources. The ICCF Group has facilitated the establishment of parliamentary conservation caucuses in ten countries and is collaborating with each of these coalitions of policymakers to strengthen governance across several key natural resource sectors. The ICCF Group coordinates high-level political engagement with expertise on conservation strategies and solutions; facilitates interactions between U.S. and international policymakers, conservation organizations, and government agencies; and seeks to leverage and integrate resources in support of sustainable natural resource management policies. ICCF-supported caucuses enable, inform, and strengthen policymakers in their commitment to conservation. -

Report of the Parliamentary Reforms and Modernisation Committee for the Third Session of the Eleventh National Assembly Appointed on 24Th September, 2014
REPORT OF THE PARLIAMENTARY REFORMS AND MODERNISATION COMMITTEE FOR THE THIRD SESSION OF THE ELEVENTH NATIONAL ASSEMBLY APPOINTED ON 24TH SEPTEMBER, 2014 Consisting of: Mr S Katuka, MP, (Chairperson); Hon A Chikwanda, MP, Minister of Finance; Hon W M Kabimba, SC, MP, Minister of Justice; Hon E Kabanshi, MP, Minister of Local Government and Housing, Hon C K Banda, SC, MP; Deputy Chairperson of Committees of the Whole House, Ms M Lubezhi, MP; Mr P Mucheleka, MP; MR L C Bwalya, MP; Mr L J Ngoma, MP; and Mr I K Banda, MP. The composition of your Committee changed in the course of the session when Hon E Kabanshi, Minister of Local Government and Housing then was appointed to the Ministry of Community Development, Mother and Child Health. She was subsequently replaced by Hon E Chenda, Minister of Commerce then who was appointed as the new Minister of Local Government and Housing. The Honourable Mr Speaker National Assembly Parliament Building LUSAKA Sir, Your Committee has the honour to present its report for the Third Session of the Eleventh National Assembly. 2. Functions of the Committee Your Committee was guided in all its deliberations by Standing Order No. 152 which set out the functions of your Committee as set out below. 1 (i) In addition to any other work placed upon it by any Standing Orders of the Assembly, it shall be the duty of the Committee to examine and propose reform to the powers, procedures and practices, organisation and facilities of the Assembly, provided that in proposing such reforms, the Committee shall bear in mind the balance of power between the respective constitutional responsibilities, roles of the National Assembly and the Government and the duties of other House Keeping Committees. -

Registered Voters by Gender and Constituency
REGISTERED VOTERS BY GENDER AND CONSTITUENCY % OF % OF SUB % OF PROVINCIAL CONSTITUENCY NAME MALES MALES FEMALES FEMALES TOTAL TOTAL KATUBA 25,040 46.6% 28,746 53.4% 53,786 8.1% KEEMBE 23,580 48.1% 25,453 51.9% 49,033 7.4% CHISAMBA 19,289 47.5% 21,343 52.5% 40,632 6.1% CHITAMBO 11,720 44.1% 14,879 55.9% 26,599 4.0% ITEZH-ITEZHI 18,713 47.2% 20,928 52.8% 39,641 5.9% BWACHA 24,749 48.1% 26,707 51.9% 51,456 7.7% KABWE CENTRAL 31,504 47.4% 34,993 52.6% 66,497 10.0% KAPIRI MPOSHI 41,947 46.7% 47,905 53.3% 89,852 13.5% MKUSHI SOUTH 10,797 47.3% 12,017 52.7% 22,814 3.4% MKUSHI NORTH 26,983 49.5% 27,504 50.5% 54,487 8.2% MUMBWA 23,494 47.9% 25,545 52.1% 49,039 7.4% NANGOMA 12,487 47.4% 13,864 52.6% 26,351 4.0% LUFUBU 5,491 48.1% 5,920 51.9% 11,411 1.7% MUCHINGA 10,072 49.7% 10,200 50.3% 20,272 3.0% SERENJE 14,415 48.5% 15,313 51.5% 29,728 4.5% MWEMBEZHI 16,756 47.9% 18,246 52.1% 35,002 5.3% 317,037 47.6% 349,563 52.4% 666,600 100.0% % OF % OF SUB % OF PROVINCIAL CONSTITUENCY NAME MALES MALES FEMALES FEMALES TOTAL TOTAL CHILILABOMBWE 28,058 51.1% 26,835 48.9% 54,893 5.4% CHINGOLA 34,695 49.7% 35,098 50.3% 69,793 6.8% NCHANGA 23,622 50.0% 23,654 50.0% 47,276 4.6% KALULUSHI 32,683 50.1% 32,614 49.9% 65,297 6.4% CHIMWEMWE 29,370 48.7% 30,953 51.3% 60,323 5.9% KAMFINSA 24,282 51.1% 23,214 48.9% 47,496 4.6% KWACHA 31,637 49.3% 32,508 50.7% 64,145 6.3% NKANA 27,595 51.9% 25,562 48.1% 53,157 5.2% WUSAKILE 23,206 50.5% 22,787 49.5% 45,993 4.5% LUANSHYA 26,658 49.5% 27,225 50.5% 53,883 5.3% ROAN 15,921 50.1% 15,880 49.9% 31,801 3.1% LUFWANYAMA 18,023 50.2% -

Members of the Northern Rhodesia Legislative Council and National Assembly of Zambia, 1924-2021
NATIONAL ASSEMBLY OF ZAMBIA Parliament Buildings P.O Box 31299 Lusaka www.parliament.gov.zm MEMBERS OF THE NORTHERN RHODESIA LEGISLATIVE COUNCIL AND NATIONAL ASSEMBLY OF ZAMBIA, 1924-2021 FIRST EDITION, 2021 TABLE OF CONTENTS FOREWORD ................................................................................................................................................ 3 PREFACE ..................................................................................................................................................... 4 ACKNOWLEDGEMENTS .......................................................................................................................... 5 ABBREVIATIONS ...................................................................................................................................... 7 INTRODUCTION ........................................................................................................................................ 9 PART A: MEMBERS OF THE LEGISLATIVE COUNCIL, 1924 - 1964 ............................................... 10 PRIME MINISTERS OF THE FEDERATION OF RHODESIA .......................................................... 12 GOVERNORS OF NORTHERN RHODESIA AND PRESIDING OFFICERS OF THE LEGISTRATIVE COUNCIL (LEGICO) ............................................................................................... 13 SPEAKERS OF THE LEGISTRATIVE COUNCIL (LEGICO) - 1948 TO 1964 ................................. 16 DEPUTY SPEAKERS OF THE LEGICO 1948 TO 1964 .................................................................... -

Chiefdoms/Chiefs in Zambia
CHIEFDOMS/CHIEFS IN ZAMBIA 1. CENTRAL PROVINCE A. Chibombo District Tribe 1 HRH Chief Chitanda Lenje People 2 HRH Chieftainess Mungule Lenje People 3 HRH Chief Liteta Lenje People B. Chisamba District 1 HRH Chief Chamuka Lenje People C. Kapiri Mposhi District 1 HRH Senior Chief Chipepo Lenje People 2 HRH Chief Mukonchi Swaka People 3 HRH Chief Nkole Swaka People D. Ngabwe District 1 HRH Chief Ngabwe Lima/Lenje People 2 HRH Chief Mukubwe Lima/Lenje People E. Mkushi District 1 HRHChief Chitina Swaka People 2 HRH Chief Shaibila Lala People 3 HRH Chief Mulungwe Lala People F. Luano District 1 HRH Senior Chief Mboroma Lala People 2 HRH Chief Chembe Lala People 3 HRH Chief Chikupili Swaka People 4 HRH Chief Kanyesha Lala People 5 HRHChief Kaundula Lala People 6 HRH Chief Mboshya Lala People G. Mumbwa District 1 HRH Chief Chibuluma Kaonde/Ila People 2 HRH Chieftainess Kabulwebulwe Nkoya People 3 HRH Chief Kaindu Kaonde People 4 HRH Chief Moono Ila People 5 HRH Chief Mulendema Ila People 6 HRH Chief Mumba Kaonde People H. Serenje District 1 HRH Senior Chief Muchinda Lala People 2 HRH Chief Kabamba Lala People 3 HRh Chief Chisomo Lala People 4 HRH Chief Mailo Lala People 5 HRH Chieftainess Serenje Lala People 6 HRH Chief Chibale Lala People I. Chitambo District 1 HRH Chief Chitambo Lala People 2 HRH Chief Muchinka Lala People J. Itezhi Tezhi District 1 HRH Chieftainess Muwezwa Ila People 2 HRH Chief Chilyabufu Ila People 3 HRH Chief Musungwa Ila People 4 HRH Chief Shezongo Ila People 5 HRH Chief Shimbizhi Ila People 6 HRH Chief Kaingu Ila People K.