Single-Chromosome Transcriptional Profiling Reveals Chromosomal Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Folic Acid and Its Receptors Jacqueline Spreadbury Governors State University
Governors State University OPUS Open Portal to University Scholarship All Capstone Projects Student Capstone Projects Spring 2013 Folic Acid and Its Receptors Jacqueline Spreadbury Governors State University Follow this and additional works at: http://opus.govst.edu/capstones Part of the Analytical Chemistry Commons Recommended Citation Spreadbury, Jacqueline, "Folic Acid and Its Receptors" (2013). All Capstone Projects. 8. http://opus.govst.edu/capstones/8 For more information about the academic degree, extended learning, and certificate programs of Governors State University, go to http://www.govst.edu/Academics/Degree_Programs_and_Certifications/ Visit the Governors State Analytical Chemistry Department This Project Summary is brought to you for free and open access by the Student Capstone Projects at OPUS Open Portal to University Scholarship. It has been accepted for inclusion in All Capstone Projects by an authorized administrator of OPUS Open Portal to University Scholarship. For more information, please contact [email protected]. Jacqueline Spreadbury Graduate Literary Review Project Spring Semester 2013 Folic Acid and its Receptors Overview of Folic Acid Folic acid, also known as folate or vitamin B9, is essential for various functions in the human body and life as we know it. Folate is the compound that occurs naturally in food, and folic acid is the synthetic form of this vitamin (1). Chemically speaking, folic acid has the hydrogen (H+) attached to the compound whereas folate is the conjugate, having lost the hydrogen (H+) (1). In the discussion below, folic acid and folate will be used interchangeably. The human body requires about 400 micrograms of folic acid daily, but cannot create folic acid on its own; instead the human diet must take in folate on a daily basis (2). -

Status of Dhps and Dhfr Genes of Plasmodium Falciparum in Colombia Before Artemisinin Based Treatment Policy
Status of dhps and dhfr genes of Plasmodium falciparum in Colombia before artemisinin based treatment policy ARTÍCULO ORIGINAL Status of dhps and dhfr genes of Plasmodium falciparum in Colombia before artemisinin based treatment policy Estado de los genes dhps y dhfr de Plasmodium falciparum en Colombia antes de la recomendación de tratamiento basado en artemisinina Andrés Villa1†, Jaime Carmona-Fonseca1, Agustín Benito2, Alonso Martínez3, Amanda Maestre1 Abstract Introduction: Surveillance of the genetic characteristics of dhps and dhfr can be useful to outline guidelines for application of intermittent preven- tive therapy in Northwest Colombia and to define the future use of antifolates in artemisinin-based combination therapy schemes. Objective: To evaluate the frequency of mutations in dhps and dhfr and to characterize parasite populations using msp-1, msp-2 and glurp in historic samples before artemisinin-based therapy was implemented in the country. Methods: A controlled clinical study was carried out on randomly selected Plasmodium falciparum infected volunteers of Northwest Colombia (Turbo and Zaragoza). A sample size of 25 subjects per region was calculated. Treatment efficacy to antifolates was assessed. Molecular analyses included P. falcipa- rum genotypes by msp-1, msp-2 and glurp and evaluation of the status of codons 16, 51, 59, 108 and 164 of dhfr and 436, 437, 540, 581 and 613 of dhps. Results: In total 78 subjects were recruited. A maximum number of 4 genotypes were detected by msp-1, msp-2 and glurp. Codons 16, 59 and 164 of the dhfr gene exhibited the wild-type form, while codons 51 and 108 were mutant. -

Snps Detection in DHPS-WDR83 Overlapping Genes Mapping On
Zambonelli et al. BMC Genetics 2013, 14:99 http://www.biomedcentral.com/1471-2156/14/99 RESEARCH ARTICLE Open Access SNPs detection in DHPS-WDR83 overlapping genes mapping on porcine chromosome 2 in a QTL region for meat pH Paolo Zambonelli1*, Roberta Davoli1, Mila Bigi1, Silvia Braglia1, Luigi Francesco De Paolis1, Luca Buttazzoni2,3, Maurizio Gallo3 and Vincenzo Russo1 Abstract Background: The pH is an important parameter influencing technological quality of pig meat, a trait affected by environmental and genetic factors. Several quantitative trait loci associated to meat pH are described on PigQTL database but only two genes influencing this parameter have been so far detected: Ryanodine receptor 1 and Protein kinase, AMP-activated, gamma 3 non-catalytic subunit. To search for genes influencing meat pH we analyzed genomic regions with quantitative effect on this trait in order to detect SNPs to use for an association study. Results: The expressed sequences mapping on porcine chromosomes 1, 2, 3 in regions associated to pork pH were searched in silico to find SNPs. 356 out of 617 detected SNPs were used to genotype Italian Large White pigs and to perform an association analysis with meat pH values recorded in semimembranosus muscle at about 1 hour (pH1) and 24 hours (pHu) post mortem. The results of the analysis showed that 5 markers mapping on chromosomes 1 or 3 were associated with pH1 and 10 markers mapping on chromosomes 1 or 2 were associated with pHu. After False Discovery Rate correction only one SNP mapping on chromosome 2 was confirmed to be associated to pHu. -

Emerging Role of ODC1 in Neurodevelopmental Disorders and Brain Development
G C A T T A C G G C A T genes Article Emerging Role of ODC1 in Neurodevelopmental Disorders and Brain Development Jeremy W. Prokop 1,2,3,*, Caleb P. Bupp 1,4 , Austin Frisch 1, Stephanie M. Bilinovich 1 , Daniel B. Campbell 1,3,5, Daniel Vogt 1,3,5, Chad R. Schultz 1, Katie L. Uhl 1, Elizabeth VanSickle 4, Surender Rajasekaran 1,6,7 and André S. Bachmann 1,* 1 Department of Pediatrics and Human Development, Michigan State University, Grand Rapids, MI 49503, USA; [email protected] (C.P.B.); [email protected] (A.F.); [email protected] (S.M.B.); [email protected] (D.B.C.); [email protected] (D.V.); [email protected] (C.R.S.); [email protected] (K.L.U.); [email protected] (S.R.) 2 Department of Pharmacology and Toxicology, Michigan State University, East Lansing, MI 48824, USA 3 Center for Research in Autism, Intellectual, and Other Neurodevelopmental Disabilities, Michigan State University, East Lansing, MI 48824, USA 4 Spectrum Health Medical Genetics, Grand Rapids, MI 49503, USA; [email protected] 5 Neuroscience Program, Michigan State University, East Lansing, MI 48824, USA 6 Pediatric Intensive Care Unit, Helen DeVos Children’s Hospital, Grand Rapids, MI 49503, USA 7 Office of Research, Spectrum Health, Grand Rapids, MI 49503, USA * Correspondence: [email protected] (J.W.P.); [email protected] (A.S.B.) Abstract: Ornithine decarboxylase 1 (ODC1 gene) has been linked through gain-of-function variants Citation: Prokop, J.W.; Bupp, C.P.; to a rare disease featuring developmental delay, alopecia, macrocephaly, and structural brain anoma- Frisch, A.; Bilinovich, S.M.; Campbell, lies. -

Systematically Profiling the Expression of Eif3 Subunits in Glioma Reveals
Chai et al. Cancer Cell Int (2019) 19:155 https://doi.org/10.1186/s12935-019-0867-1 Cancer Cell International PRIMARY RESEARCH Open Access Systematically profling the expression of eIF3 subunits in glioma reveals the expression of eIF3i has prognostic value in IDH-mutant lower grade glioma Rui‑Chao Chai1,4,6†, Ning Wang2†, Yu‑Zhou Chang3, Ke‑Nan Zhang1,6, Jing‑Jun Li1,6, Jun‑Jie Niu5, Fan Wu1,6*, Yu‑Qing Liu1,6* and Yong‑Zhi Wang1,3,4,6* Abstract Background: Abnormal expression of the eukaryotic initiation factor 3 (eIF3) subunits plays critical roles in tumo‑ rigenesis and progression, and also has potential prognostic value in cancers. However, the expression and clinical implications of eIF3 subunits in glioma remain unknown. Methods: Expression data of eIF3 for patients with gliomas were obtained from the Chinese Glioma Genome Atlas (CGGA) (n 272) and The Cancer Genome Atlas (TCGA) (n 595). Cox regression, the receiver operating characteristic (ROC) curves= and Kaplan–Meier analysis were used to study= the prognostic value. Gene oncology (GO) and gene set enrichment analysis (GSEA) were utilized for functional prediction. Results: In both the CGGA and TCGA datasets, the expression levels of eIF3d, eIF3e, eIF3f, eIF3h and eIF3l highly were associated with the IDH mutant status of gliomas. The expression of eIF3b, eIF3i, eIF3k and eIF3m was increased with the tumor grade, and was associated with poorer overall survival [All Hazard ratio (HR) > 1 and P < 0.05]. By contrast, the expression of eIF3a and eIF3l was decreased in higher grade gliomas and was associated with better overall sur‑ vival (Both HR < 1 and P < 0.05). -

Translation Initiation Factor Eif3h Targets Specific Transcripts To
Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis Avik Choudhuria,b, Umadas Maitraa,1, and Todd Evansb,1 aDepartment of Developmental and Molecular Biology, Albert Einstein College of Medicine of Yeshiva University, Bronx, NY 10461; and bDepartment of Surgery, Weill Cornell Medical College, New York, NY 10065 Edited by Igor B. Dawid, The Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, and approved April 30, 2013 (received for review February 14, 2013) Eukaryotic translation initiation factor 3 (eIF3) plays a central role eukaryotes. These—eIF3d, eIF3e, eIF3f, eIF3h, eIF3j, eIF3k, in translation initiation and consists of five core (conserved) sub- eIF3l, and eIF3m—were designated “non-core” subunits (4). units present in both budding yeast and higher eukaryotes. Higher In contrast to the budding yeast, the genome of the fission eukaryotic eIF3 contains additional (noncore or nonconserved) yeast Schizosaccharomyces pombe contains structural homologs subunits of poorly defined function, including sub-unit h (eIF3h), of at least five noncore (nonconserved) eIF3 subunits—eIF3d, which in zebrafish is encoded by two distinct genes (eif3ha and eIF3e, eIF3f, eIF3h, and eIF3m. The gene encoding eIF3f is eif3hb). Previously we showed that eif3ha encodes the predominant essential for growth, whereas eIF3d, eIF3e, and eIF3h are dis- isoform during zebrafish embryogenesis and that depletion of this pensable for growth and viability (5–11). However, deleted factor causes defects in the development of the brain and eyes. To strains show specific phenotypes including defects in meiosis/ investigate the molecular mechanism governing this regulation, we sporulation (6, 9, 11). -

Identification of Differentially Expressed Genes and Pathways in Mice Exposed to Mixed Field Neutron/Photon Radiation Constantinos G
Broustas et al. BMC Genomics (2018) 19:504 https://doi.org/10.1186/s12864-018-4884-6 RESEARCHARTICLE Open Access Identification of differentially expressed genes and pathways in mice exposed to mixed field neutron/photon radiation Constantinos G. Broustas1* , Andrew D. Harken2, Guy Garty2 and Sally A. Amundson1 Abstract Background: Radiation exposure due to the detonation of an improvised nuclear device remains a major security concern. Radiation from such a device involves a combination of photons and neutrons. Although photons will make the greater contribution to the total dose, neutrons will certainly have an impact on the severity of the exposure as they have high relative biological effectiveness. Results: We investigated the gene expression signatures in the blood of mice exposed to 3 Gy x-rays, 0.75 Gy of neutrons, or to mixed field photon/neutron with the neutron fraction contributing 5, 15%, or 25% of a total 3 Gy radiation dose. Gene ontology and pathway analysis revealed that genes involved in protein ubiquitination pathways were significantly overrepresented in all radiation doses and qualities. On the other hand, eukaryotic initiation factor 2 (EIF2) signaling pathway was identified as one of the top 10 ranked canonical pathways in neutron, but not pure x-ray, exposures. In addition, the related mTOR and regulation of EIF4/p70S6K pathways were also significantly underrepresented in the exposures with a neutron component, but not in x-ray radiation. The majority of the changed genes in these pathways belonged to the ribosome biogenesis and translation machinery and included several translation initiation factors (e.g. Eif2ak4, Eif3f), as well as 40S and 60S ribosomal subunits (e.g. -

1 1 2 Pharmacological Dimerization and Activation of the Exchange
1 2 3 Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the 4 integrated stress response 5 6 7 *Carmela Sidrauski1,2, *Jordan C. Tsai1,2, Martin Kampmann2,3, Brian R. Hearn4, Punitha 8 Vedantham4, Priyadarshini Jaishankar4 , Masaaki Sokabe5, Aaron S. Mendez1,2, Billy W. 9 Newton6, Edward L. Tang6.7, Erik Verschueren6, Jeffrey R. Johnson6,7, Nevan J. Krogan6,7,, 10 Christopher S. Fraser5, Jonathan S. Weissman2,3, Adam R. Renslo4, and Peter Walter 1,2 11 12 1Department of Biochemistry and Biophysics, University of California, San Francisco, United 13 States 14 2Howard Hughes Medical Institute, University of California, San Francisco, United States 15 3Department of Cellular and Molecular Pharmacology, University of California, San Francisco, 16 United States 17 4Department of Pharmaceutical Chemistry and the Small Molecule Discovery Center, University 18 of California at San Francisco, United States 19 5Department of Molecular and Cellular Biology, College of Biological Sciences, University of 20 California, Davis, United States 21 6QB3, California Institute for Quantitative Biosciences, University of California, San Francisco, 22 United States 23 7Gladstone Institutes, San Francisco, United States 24 25 * Both authors contributed equally to this work 26 27 28 Abstract 29 30 The general translation initiation factor eIF2 is a major translational control point. Multiple 31 signaling pathways in the integrated stress response phosphorylate eIF2 serine-51, inhibiting 32 nucleotide exchange by eIF2B. ISRIB, a potent drug-like small molecule, renders cells 33 insensitive to eIF2α phosphorylation and enhances cognitive function in rodents by blocking 34 long-term depression. ISRIB was identified in a phenotypic cell-based screen, and its mechanism 35 of action remained unknown. -

Relevance of Translation Initiation in Diffuse Glioma Biology and Its
cells Review Relevance of Translation Initiation in Diffuse Glioma Biology and its Therapeutic Potential Digregorio Marina 1, Lombard Arnaud 1,2, Lumapat Paul Noel 1, Scholtes Felix 1,2, Rogister Bernard 1,3 and Coppieters Natacha 1,* 1 Laboratory of Nervous System Disorders and Therapy, GIGA-Neurosciences Research Centre, University of Liège, 4000 Liège, Belgium; [email protected] (D.M.); [email protected] (L.A.); [email protected] (L.P.N.); [email protected] (S.F.); [email protected] (R.B.) 2 Department of Neurosurgery, CHU of Liège, 4000 Liège, Belgium 3 Department of Neurology, CHU of Liège, 4000 Liège, Belgium * Correspondence: [email protected] Received: 18 October 2019; Accepted: 26 November 2019; Published: 29 November 2019 Abstract: Cancer cells are continually exposed to environmental stressors forcing them to adapt their protein production to survive. The translational machinery can be recruited by malignant cells to synthesize proteins required to promote their survival, even in times of high physiological and pathological stress. This phenomenon has been described in several cancers including in gliomas. Abnormal regulation of translation has encouraged the development of new therapeutics targeting the protein synthesis pathway. This approach could be meaningful for glioma given the fact that the median survival following diagnosis of the highest grade of glioma remains short despite current therapy. The identification of new targets for the development of novel therapeutics is therefore needed in order to improve this devastating overall survival rate. This review discusses current literature on translation in gliomas with a focus on the initiation step covering both the cap-dependent and cap-independent modes of initiation. -

Estrogenic Endocrine Disrupting Chemicals Influencing NRF1 Regulated Gene Networks in the Development of Complex Human Brain
Florida International University FIU Digital Commons All Faculty 12-13-2016 Estrogenic Endocrine Disrupting Chemicals Influencing NRF1 Regulated Gene Networks in the Development of Complex Human Brain Diseases Mark Preciados Department of Environmental & Occupational Health, Florida International University, [email protected] Changwon Yoo Department of Biostatistics, Florida International University, [email protected] Deodutta Roy Department of Environmental & Occupational Health, Florida International University, [email protected] Follow this and additional works at: https://digitalcommons.fiu.edu/all_faculty Recommended Citation Preciados, Mark; Yoo, Changwon; and Roy, Deodutta, "Estrogenic Endocrine Disrupting Chemicals Influencing NRF1 Regulated Gene Networks in the Development of Complex Human Brain Diseases" (2016). All Faculty. 180. https://digitalcommons.fiu.edu/all_faculty/180 This work is brought to you for free and open access by FIU Digital Commons. It has been accepted for inclusion in All Faculty by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. International Journal of Molecular Sciences Review Estrogenic Endocrine Disrupting Chemicals Influencing NRF1 Regulated Gene Networks in the Development of Complex Human Brain Diseases Mark Preciados 1, Changwon Yoo 2 and Deodutta Roy 1,* 1 Department of Environmental & Occupational Health, Florida International University, Miami, FL 33199, USA; mprec001@fiu.edu 2 Department of Biostatistics, Florida International University, Miami, FL 33199, USA; cyoo@fiu.edu * Correspondence: droy@fiu.edu; Tel.: +1-305-348-1694; Fax: +1-305-348-4901 Academic Editor: Paul B. Tchounwou Received: 25 October 2016; Accepted: 29 November 2016; Published: 13 December 2016 Abstract: During the development of an individual from a single cell to prenatal stages to adolescence to adulthood and through the complete life span, humans are exposed to countless environmental and stochastic factors, including estrogenic endocrine disrupting chemicals. -
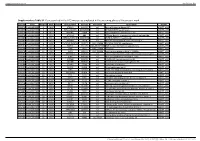
Supplementary Table S1. Genes Printed in the HC5 Microarray Employed in the Screening Phase of the Present Work
Supplementary material Ann Rheum Dis Supplementary Table S1. Genes printed in the HC5 microarray employed in the screening phase of the present work. CloneID Plate Position Well Length GeneSymbol GeneID Accession Description Vector 692672 HsxXG013989 2 B01 STK32A 202374 null serine/threonine kinase 32A pANT7_cGST 692675 HsxXG013989 3 C01 RPS10-NUDT3 100529239 null RPS10-NUDT3 readthrough pANT7_cGST 692678 HsxXG013989 4 D01 SPATA6L 55064 null spermatogenesis associated 6-like pANT7_cGST 692679 HsxXG013989 5 E01 ATP1A4 480 null ATPase, Na+/K+ transporting, alpha 4 polypeptide pANT7_cGST 692689 HsxXG013989 6 F01 ZNF816-ZNF321P 100529240 null ZNF816-ZNF321P readthrough pANT7_cGST 692691 HsxXG013989 7 G01 NKAIN1 79570 null Na+/K+ transporting ATPase interacting 1 pANT7_cGST 693155 HsxXG013989 8 H01 TNFSF12-TNFSF13 407977 NM_172089 TNFSF12-TNFSF13 readthrough pANT7_cGST 693161 HsxXG013989 9 A02 RAB12 201475 NM_001025300 RAB12, member RAS oncogene family pANT7_cGST 693169 HsxXG013989 10 B02 SYN1 6853 NM_133499 synapsin I pANT7_cGST 693176 HsxXG013989 11 C02 GJD3 125111 NM_152219 gap junction protein, delta 3, 31.9kDa pANT7_cGST 693181 HsxXG013989 12 D02 CHCHD10 400916 null coiled-coil-helix-coiled-coil-helix domain containing 10 pANT7_cGST 693184 HsxXG013989 13 E02 IDNK 414328 null idnK, gluconokinase homolog (E. coli) pANT7_cGST 693187 HsxXG013989 14 F02 LYPD6B 130576 null LY6/PLAUR domain containing 6B pANT7_cGST 693189 HsxXG013989 15 G02 C8orf86 389649 null chromosome 8 open reading frame 86 pANT7_cGST 693194 HsxXG013989 16 H02 CENPQ 55166 -

Nat1 Promotes Translation of Specific Proteins That Induce Differentiation of Mouse Embryonic Stem Cells
Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells Hayami Sugiyamaa, Kazutoshi Takahashia,b, Takuya Yamamotoa,c,d, Mio Iwasakia, Megumi Naritaa, Masahiro Nakamuraa, Tim A. Randb, Masato Nakagawaa, Akira Watanabea,c,e, and Shinya Yamanakaa,b,1 aDepartment of Life Science Frontiers, Center for iPS Cell Research and Application, Kyoto University, Kyoto 606-8507, Japan; bGladstone Institute of Cardiovascular Disease, San Francisco, CA 94158; cInstitute for Integrated Cell-Material Sciences, Kyoto University, Kyoto 606-8501, Japan; dJapan Agency for Medical Research and Development-Core Research for Evolutional Science and Technology (AMED-CREST), Tokyo 100-0004, Japan; and eJapan Science and Technology Agency (JST)-CREST, Saitama 332-0012, Japan Contributed by Shinya Yamanaka, November 22, 2016 (sent for review October 18, 2016; reviewed by Katsura Asano and Keisuke Kaji) Novel APOBEC1 target 1 (Nat1) (also known as “p97,”“Dap5,” and To study the role of Nat1 in cell differentiation further, we “Eif4g2”) is a ubiquitously expressed cytoplasmic protein that is homol- generated mouse embryonic stem cells (mES cells) lacking both ogous to the C-terminal two thirds of eukaryotic translation initiation alleles of the Nat1 gene. mES cells were derived from blastocysts factor 4G (Eif4g1). We previously showed that Nat1-null mouse embry- in 1981 (11, 12) and possess two unique properties. First, ES cells onic stem cells (mES cells) are resistant to differentiation. In the current have the potential to self-renew indefinitely (maintenance). Sec- study, we found that NAT1 and eIF4G1 share many binding proteins, ond, ES cells have the potential to differentiate into all somatic such as the eukaryotic translation initiation factors eIF3 and eIF4A and and germ cell types (pluripotency) that make up the body.