Additional File 1: SDS-PAGE Fractionation with Silver Staining of Bone Marrow Supernatant from Six Β- Thalassemia/Hb E Patients (P) and Four Donors (D)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Nitroxide-Mediated Polymerization
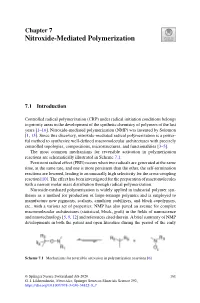
Chapter 7 Nitroxide-Mediated Polymerization 7.1 Introduction Controlled radical polymerization (CRP) under radical initiation conditions belongs to priority areas in the development of the synthetic chemistry of polymers of the last years [1–16]. Nitroxide-mediated polymerization (NMP) was invented by Solomon [1, 13]. Since this discovery, nitroxide-mediated radical polymerization is a power- ful method to synthesize well-defined macromolecular architectures with precisely controlled topologies, compositions, microstructures, and functionalities [3–5]. The most common mechanisms for reversible activation in polymerization reactions are schematically illustrated in Scheme 7.1. Persistent radical effect (PRE) occurs when two radicals are generated at the same time, at the same rate, and one is more persistent than the other, the self-termination reactions are lowered, leading to an unusually high selectivity for the cross-coupling reaction [10]. The effect has been investigated for the preparation of macromolecules with a narrow molar mass distribution through radical polymerization. Nitroxide-mediated polymerization is widely applied in industrial polymer syn- theses as a method for production of large-tonnage polymers and is employed to manufacture new pigments, sealants, emulsion stabilizers, and block copolymers, etc., with a various set of properties. NMP has also paved an avenue for complex macromolecular architectures (statistical, block, graft) in the fields of nanoscience and nanotechnology [5, 9, 12] and references cited therein. A brief summary of NMP developments in both the patent and open literature during the period of the early Scheme 7.1 Mechanisms for reversible activation in polymerization reactions [6] © Springer Nature Switzerland AG 2020 161 G. I. Likhtenshtein, Nitroxides, Springer Series in Materials Science 292, https://doi.org/10.1007/978-3-030-34822-9_7 162 7 Nitroxide-Mediated Polymerization 1980–2000 was presented in [11]. -

SPATA33 Localizes Calcineurin to the Mitochondria and Regulates Sperm Motility in Mice
SPATA33 localizes calcineurin to the mitochondria and regulates sperm motility in mice Haruhiko Miyataa, Seiya Ouraa,b, Akane Morohoshia,c, Keisuke Shimadaa, Daisuke Mashikoa,1, Yuki Oyamaa,b, Yuki Kanedaa,b, Takafumi Matsumuraa,2, Ferheen Abbasia,3, and Masahito Ikawaa,b,c,d,4 aResearch Institute for Microbial Diseases, Osaka University, Osaka 5650871, Japan; bGraduate School of Pharmaceutical Sciences, Osaka University, Osaka 5650871, Japan; cGraduate School of Medicine, Osaka University, Osaka 5650871, Japan; and dThe Institute of Medical Science, The University of Tokyo, Tokyo 1088639, Japan Edited by Mariana F. Wolfner, Cornell University, Ithaca, NY, and approved July 27, 2021 (received for review April 8, 2021) Calcineurin is a calcium-dependent phosphatase that plays roles in calcineurin can be a target for reversible and rapidly acting male a variety of biological processes including immune responses. In sper- contraceptives (5). However, it is challenging to develop molecules matozoa, there is a testis-enriched calcineurin composed of PPP3CC and that specifically inhibit sperm calcineurin and not somatic calci- PPP3R2 (sperm calcineurin) that is essential for sperm motility and male neurin because of sequence similarities (82% amino acid identity fertility. Because sperm calcineurin has been proposed as a target for between human PPP3CA and PPP3CC and 85% amino acid reversible male contraceptives, identifying proteins that interact with identity between human PPP3R1 and PPP3R2). Therefore, identi- sperm calcineurin widens the choice for developing specific inhibitors. fying proteins that interact with sperm calcineurin widens the choice Here, by screening the calcineurin-interacting PxIxIT consensus motif of inhibitors that target the sperm calcineurin pathway. in silico and analyzing the function of candidate proteins through the The PxIxIT motif is a conserved sequence found in generation of gene-modified mice, we discovered that SPATA33 inter- calcineurin-binding proteins (8, 9). -

Can Alzheimer's Disease Shed Light on the DNA As ''Data'' Versus DNA
Commentary Annals of Genetics and Genetic Disorders Published: 21 Jun, 2018 Can Alzheimer’s Disease Shed Light on the DNA as ‘’Data’’ versus DNA as a ‘’Program’’ Paradigm? Bajic Vladan1*, Panagiotis Athanasios2 and Misic Natasa3 1Department of Radiobiology and Molecular Genetics, University of Belgrade, Serbia 2Department of Biotechnology, Agricultural University of Athens, Greece 3Department of Biotechnology, Research and Development Institute Lola Ltd, Serbia Commentary For 100 years it was established that Alzheimer’s disease can occur early (before 50) and late (after 65), but only in the 1970 the genetic makeup has been established for this disease [1]. Scientists pinpointed the cause of AD on two chromosomes, chromosome 21 and 14, finding that genes for APP and Presenilin are connected to how the amyloid was processed (the famous amyloid that Dr Alzheimer reported to be seen in the brain of the first patient, Auguste D). These findings and the notion that aneuploidy of chromosome 21 in Down syndrome leads to early AD suggested that etiological factor was found, even though that only 3% to 5% of all AD patients have mutations in these genes. AD cases of 95% are labeled as Sporadic AD (SAD) [1]. The genome project and later new technologies that utilize genetic screening and analysis opened a field of investigation to find the ‘’other‘’ risk genes that are in the base of SAD. This paradigm was and has been led by the viewpoint that DNA is a program. This view was established through the workings of Dr Ernst Mayer, 1961 [2] as still pursued today. On the other hand Henry Atlan, 2011 suggested that DNA is a data center and the program is utilized from what he called the ‘’complexity’’ of a cell [3]. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Genetic Basis of Dupuytren's Disease Gloria Sue Yale School of Medicine, [email protected]
Yale University EliScholar – A Digital Platform for Scholarly Publishing at Yale Yale Medicine Thesis Digital Library School of Medicine January 2014 The Genetic Basis Of Dupuytren's Disease Gloria Sue Yale School of Medicine, [email protected] Follow this and additional works at: http://elischolar.library.yale.edu/ymtdl Recommended Citation Sue, Gloria, "The Genetic Basis Of Dupuytren's Disease" (2014). Yale Medicine Thesis Digital Library. 1926. http://elischolar.library.yale.edu/ymtdl/1926 This Open Access Thesis is brought to you for free and open access by the School of Medicine at EliScholar – A Digital Platform for Scholarly Publishing at Yale. It has been accepted for inclusion in Yale Medicine Thesis Digital Library by an authorized administrator of EliScholar – A Digital Platform for Scholarly Publishing at Yale. For more information, please contact [email protected]. The Genetic Basis of Dupuytren’s Disease A Thesis Submitted to the Yale University School of Medicine In Partial Fulfillment of the Requirements for the Degree of Doctor of Medicine by Gloria R. Sue 2014 THE GENETIC BASIS OF DUPUYTREN’S DISEASE. Gloria R. Sue, Deepak Narayan. Section of Plastic and Reconstructive Surgery, Department of Surgery, Yale University School of Medicine, New Haven, CT. Dupuytren’s disease is a common heritable connective tissue disorder of poorly understood etiology. It is thought that oxidative stress pathways may play a critical role in the development of Dupuytren’s disease, given the various disease associations that have been observed. We sought to sequence the mitochondrial and nuclear genomes of patients affected with Dupuytren’s disease using next-generation sequencing technology to potentially identify genes of potential pathogenetic interest. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Ribonucleotide Reductase Subunit M2B Deficiency Leads To
Am J Transl Res 2018;10(11):3635-3649 www.ajtr.org /ISSN:1943-8141/AJTR0084684 Original Article Ribonucleotide reductase subunit M2B deficiency leads to mitochondrial permeability transition pore opening and is associated with aggressive clinicopathologic manifestations of breast cancer Lijun Xue1*, Xiyong Liu2,6*, Qinchuan Wang3,4, Charlie Q Liu3, Yunru Chen3, Wei Jia5, Ronhong Hsie6, Yifan Chen8, Frank Luh2,6, Shu Zheng7, Yun Yen2,6,8 1Department of Pathology, Loma Linda University Medical Center, Loma Linda, CA 92354, USA; 2Sino-American Cancer Foundation, California Cancer Institute, Temple, CA 91780, USA; 3Department of Molecular Pharmacology, Beckman Research Institute, City of Hope Comprehensive Cancer Center, Duarte, CA 91010, USA; 4Surgical Oncology, Sir Runrun Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China; 5Cancer Epidemiology Program, University of Hawaii Cancer Center, Honolulu, HI 96813, USA; 6TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei, Taiwan, ROC; 7Cancer Institute, Zhejiang University, Hangzhou 310009, Zhejiang, China; 8PhD Program of Cancer Biology and Drug Discovery, Taipei Medical University, Taipei, Taiwan, ROC. *Co-first authors. Received August 27, 2018; Accepted October 19, 2018; Epub November 15, 2018; Published November 30, 2018 Abstract: Ribonucleotide reductase small subunit M2B (RRM2B) plays an essential role in maintaining mitochon- drial homeostasis. Mitochondrial permeability transition pore (MPTP) is a key regulator of mitochondrial homeo- stasis. MPTP contributes to cell death and is crucial in cancer progression. RRM2B’s relation to MPTP is not well known, and the role of RRM2B in cancer progression is controversial. Here, our aim was to study the role of RRM2B in regulating MPTP and the association between RRM2B and clinicopathological manifestations in breast cancer. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Mitochondrial Hepatopathies Etiology and Genetics the Hepatocyte Mitochondrion Can Function Both As a Cause and As a Target of Liver Injury
Mitochondrial Hepatopathies Etiology and Genetics The hepatocyte mitochondrion can function both as a cause and as a target of liver injury. Most mitochondrial hepatopathies involve defects in the mitochondrial respiratory chain enzyme complexes (Figure 1). Resultant dysfunction of mitochondria yields deficient oxidative phosphorylation (OXPHOS), increased generation of reactive oxygen species (ROS), accumulation of hepatocyte lipid, impairment of other metabolic pathways and activation of both apoptotic and necrotic pathways of cellular death. Figure 1: Since the mitochondria are under dual control of nuclear DNA and mitochondrial DNA (mtDNA), mutations in genes of both classes have been associated with inherited mitochondrial myopathies, encephalopathies, and hepatopathies. Autosomal nuclear gene defects affect a variety of mitochondrial processes such as protein assembly, mtDNA synthesis and replication (e.g., deoxyguanosine kinase [dGUOK]) and DNA polymerase gamma [POLG]), and transport of nucleotides or metals. MPV17 (function unknown) and RRM2B (encoding the cytosolic p53-inducible ribonucleotide reductase small subunit) are two genes recently identified as also causing mtDNA depletion syndrome and liver failure, as has TWINKLE, TRMU, and SUCLG1. Most children with mitochondrial hepatopathies have identified or presumed mutations in these nuclear genes, rather than mtDNA genes. A classification of primary mitochondrial hepatopathies involving energy metabolism is presented in Table 1. Drug interference with mtDNA replication is now recognized as a cause of acquired mtDNA depletion that can result in liver failure, lactic acidosis, and myopathy in human immunodeficiency virus infected patients and, previously, in hepatitis B virus patients treated with nucleoside reverse transcriptase inhibitors. Current estimates suggest a minimum prevalence of all mitochondrial diseases of 11.5 cases per 100,000 individuals, or 1 in 8500 of the general population. -

Original Article a Database and Functional Annotation of NF-Κb Target Genes
Int J Clin Exp Med 2016;9(5):7986-7995 www.ijcem.com /ISSN:1940-5901/IJCEM0019172 Original Article A database and functional annotation of NF-κB target genes Yang Yang, Jian Wu, Jinke Wang The State Key Laboratory of Bioelectronics, Southeast University, Nanjing 210096, People’s Republic of China Received November 4, 2015; Accepted February 10, 2016; Epub May 15, 2016; Published May 30, 2016 Abstract: Backgrounds: The previous studies show that the transcription factor NF-κB always be induced by many inducers, and can regulate the expressions of many genes. The aim of the present study is to explore the database and functional annotation of NF-κB target genes. Methods: In this study, we manually collected the most complete listing of all NF-κB target genes identified to date, including the NF-κB microRNA target genes and built the database of NF-κB target genes with the detailed information of each target gene and annotated it by DAVID tools. Results: The NF-κB target genes database was established (http://tfdb.seu.edu.cn/nfkb/). The collected data confirmed that NF-κB maintains multitudinous biological functions and possesses the considerable complexity and diversity in regulation the expression of corresponding target genes set. The data showed that the NF-κB was a central regula- tor of the stress response, immune response and cellular metabolic processes. NF-κB involved in bone disease, immunological disease and cardiovascular disease, various cancers and nervous disease. NF-κB can modulate the expression activity of other transcriptional factors. Inhibition of IKK and IκBα phosphorylation, the decrease of nuclear translocation of p65 and the reduction of intracellular glutathione level determined the up-regulation or down-regulation of expression of NF-κB target genes. -

Identification of Transcriptional Mechanisms Downstream of Nf1 Gene Defeciency in Malignant Peripheral Nerve Sheath Tumors Daochun Sun Wayne State University
Wayne State University DigitalCommons@WayneState Wayne State University Dissertations 1-1-2012 Identification of transcriptional mechanisms downstream of nf1 gene defeciency in malignant peripheral nerve sheath tumors Daochun Sun Wayne State University, Follow this and additional works at: http://digitalcommons.wayne.edu/oa_dissertations Recommended Citation Sun, Daochun, "Identification of transcriptional mechanisms downstream of nf1 gene defeciency in malignant peripheral nerve sheath tumors" (2012). Wayne State University Dissertations. Paper 558. This Open Access Dissertation is brought to you for free and open access by DigitalCommons@WayneState. It has been accepted for inclusion in Wayne State University Dissertations by an authorized administrator of DigitalCommons@WayneState. IDENTIFICATION OF TRANSCRIPTIONAL MECHANISMS DOWNSTREAM OF NF1 GENE DEFECIENCY IN MALIGNANT PERIPHERAL NERVE SHEATH TUMORS by DAOCHUN SUN DISSERTATION Submitted to the Graduate School of Wayne State University, Detroit, Michigan in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY 2012 MAJOR: MOLECULAR BIOLOGY AND GENETICS Approved by: _______________________________________ Advisor Date _______________________________________ _______________________________________ _______________________________________ © COPYRIGHT BY DAOCHUN SUN 2012 All Rights Reserved DEDICATION This work is dedicated to my parents and my wife Ze Zheng for their continuous support and understanding during the years of my education. I could not achieve my goal without them. ii ACKNOWLEDGMENTS I would like to express tremendous appreciation to my mentor, Dr. Michael Tainsky. His guidance and encouragement throughout this project made this dissertation come true. I would also like to thank my committee members, Dr. Raymond Mattingly and Dr. John Reiners Jr. for their sustained attention to this project during the monthly NF1 group meetings and committee meetings, Dr. -

01-11 Genetics of Alzheimer
DERLEME/REVIEW Genetics of Alzheimer’s Disease: Lessons Learned in Two Decades Alzheimer Hastalığının Genetiği: Son 20 Yılda Öğrenilen Dersler Nilüfer Ertekin Taner Mayo Klinik Florida, Nöroloji ve Nörobilim Bölümleri, Jacksonville, Florida, Amerika Birleşik Devletleri Turk Norol Derg 2010;16:1-11 ÖZET Alzheimer hastal›¤› (AH) en s›k rastlanan demans türüdür. 2010 y›l›nda tüm demanslar›n dünyada 35 milyondan fazla kifliyi etkileme- si beklenmektedir. Etkili tedaviler olmaks›z›n, bu salg›n›n 2050 y›l›nda tüm dünyada 115 milyondan fazla hasta say›s›na ulaflaca¤› he- saplanmaktad›r. Genetik çal›flmalar hastal›¤›n patofizyolojisini anlamaya yarayarak, olas› tedavi, semptom öncesi tan› ve önlemlere yol açabilir. 1990 y›l›ndan bu yana AH’›n altta yatan genetik ögesi hakk›nda önemli oranda kan›t birikmifltir. Erken bafllang›çl› ailesel AH’a yol açan otozomal dominant mutasyonlar tafl›yan üç gen, AH’›n %1’inden daha az›n› aç›klamaktad›r. Geç bafllang›çl› AH’daki genel kabul gören tek risk faktörü olan apolipoprotein ε4, bu hastal›¤›n genetik riskinin yaln›zca bir k›sm›n› aç›klar. Genetik ba¤lant› ve ilifl- ki çal›flmalar›nda birçok aday gen bölgesi bulunmas›na ra¤men, bu sonuçlar ba¤›ms›z çal›flmalarda ço¤unlukla tekrarlanamam›flt›r. Bu- nun nedeni, en az›ndan k›smen, genetik heterojenlik, düflük etkili genetik faktörler ve yetersiz güçte olan çal›flmalard›r. Yüz binlerce tekli nükleotid polimorfizmi ile binlerce kiflinin incelendi¤i genom çap›nda iliflki çal›flmalar›, AH gibi karmafl›k geneti¤e sahip hastal›k- lar›n alt›nda yatan yayg›n risk varyasyonlar›n›n bulunmas› için olas› güçlü bir yaklafl›m olarak görülmektedir.