A Comparison of the Small Ribosomal RNA Genes from the Mitochondrial DNA of the Great Apes and Humans: Sequence, Structure, Evolution, and Phylogenetic Implications
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Domain of Escherichia Coli 16S Ribosomal RNA Using Site-Directed Photoaffinity Crosslinking
Downloaded from rnajournal.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press RNA (1998), 4:1455–1466+ Cambridge University Press+ Printed in the USA+ Copyright © 1998 RNA Society+ Analysis of the conformation of the 39 major domain of Escherichia coli 16S ribosomal RNA using site-directed photoaffinity crosslinking ALEXANDRE MONTPETIT,1 CATHERINE PAYANT,1 JAMES M. NOLAN,2 and LÉA BRAKIER-GINGRAS1 1Département de Biochimie, Université de Montréal, Montréal, Québec H3T 1J4, Canada 2Department of Biochemistry, Tulane University Medical Center, New Orleans, Louisiana 70112, USA ABSTRACT The 39 major domain of Escherichia coli 16S rRNA, which occupies the head of the small ribosomal subunit, is involved in several functions of the ribosome. We have used a site-specific crosslinking procedure to gain further insights into the higher-order structure of this domain. Circularly permuted RNAs were used to introduce an azi- dophenacyl group at specific positions within the 39 major domain. Crosslinks were generated in a high-ionic strength buffer that has been used for ribosome reconstitution studies and so enables the RNA to adopt a structure recognized by ribosomal proteins. The crosslinking sites were identified by primer extension and confirmed by assessing the mobility of the crosslinked RNA lariats in denaturing polyacrylamide gels. Eight crosslinks were characterized. Among them, one crosslink demonstrates that helix 28 is proximal to the top of helix 34, and two others show that the 1337 region, located in an internal loop at the junction of helices 29, 30, 41, and 42, is proximal to the center of helix 30 and to a segment connecting helix 28 to helix 29. -

The “Tolerant Chimpanzee”—Towards the Costs and Benefits of Sociality in Female Bonobos
Behavioral The official journal of the ISBE Ecology International Society for Behavioral Ecology Behavioral Ecology (2018), 29(6), 1325–1339. doi:10.1093/beheco/ary118 Original Article The “tolerant chimpanzee”—towards the Downloaded from https://academic.oup.com/beheco/article-abstract/29/6/1325/5090310 by MPI evolutionary Anthropology user on 07 January 2019 costs and benefits of sociality in female bonobos Niina O. Nurmi,a,b, Gottfried Hohmann,b Lucas G. Goldstone,c Tobias Deschner,b and Oliver Schülkea,d aDepartment of Behavioral Ecology, JFB Institute for Zoology/Anthropology, University of Göttingen, Germany, bDepartment of Primatology, Max Planck Institute for Evolutionary Anthropology, Germany, cGraduate School of Systemic Neurosciences, Ludwig Maximilians University, Germany, and dResearch Group Social Evolution in Primates, German Primate Center, Leibniz Institute for Primate Research, Göttingen, Germany Received 7 December 2017; revised 26 July 2018; editorial decision 28 July 2018; accepted 8 August 2018; Advance Access publication 4 September 2018. Humans share an extraordinary degree of sociality with other primates, calling for comparative work into the evolutionary drivers of the variation in social engagement observed between species. Of particular interest is the contrast between the chimpanzee (Pan troglodytes) and bonobo (Pan paniscus), the latter exhibiting increased female gregariousness, more tolerant relationships, and elab- orate behavioral adaptations for conflict resolution. Here, we test predictions from 3 socioecological hypotheses regarding the evo- lution of these traits using data on wild bonobos at LuiKotale, Democratic Republic of Congo. Focusing on the behavior of co-feeding females and controlling for variation in characteristics of the feeding patch, food intake rate moderately increased while feeding effort decreased with female dominance rank, indicating that females engaged in competitive exclusion from high-quality food resources. -

Journal of School Violence
Journal of School Violence eHAWORTH® Electronic Text is provided AS IS without warranty of any kind. The Haworth Press, Inc. further disclaims all implied warranties including, without limitation, any implied warranties of merchantability or of fitness for a particular purpose. The entire risk arising out of the use of the Electronic Text remains with you. In no event shall The Haworth Press, Inc., its authors, or anyone else involved in the creation, production, or delivery of this product be liable for any damages whatsoever (including, without limitation, damages for loss of business profits, business interruption, loss of business information, or other pecuniary loss) arising out of the use of or inability to use the Electronic Text, even if The Haworth Press, Inc. has been advised of the possibility of such damages. EDITOR EDWIN R. GERLER, Jr., Professor, Counselor Education Program, College of Education, North Carolina State University, Raleigh, NC ASSOCIATE EDITORS PAMELA L. RILEY, Executive Director, National Association of Students Against Violence Everywhere (SAVE), Raleigh, NC JOANNE McDANIEL, Director, Center for the Prevention of School Violence, Raleigh, NC COLUMN EDITOR, E-SITES FOR SAFE SCHOOLS REBECCA R. REED, Ahlgren Associates, Raleigh, NC EDITORIAL BOARD DAVID P. ADAY, Jr., Department of Sociology, College of William and Mary, Williamsburg, VA RON AVI ASTOR, School of Social Work, University of Southern California, Los Angeles, CA RAMI BENBENISHTY, Paul Baerwald School of Social Work, The Hebrew University of Jerusalem, Israel ILENE R. BERSON, Department of Child and Family Studies, Louis de la Parte Florida Mental Health Institute, University of South Florida, Tampa, FL CATHERINE BLAYA-DEBARBIEUX, Universite Victor Segalen Bordeaux 2, Bordeaux Cedex, France CHERYL L. -

Unraveling the Evolutionary History of Orangutans (Genus: Pongo)- the Impact of Environmental Processes and the Genomic Basis of Adaptation
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2015 Unraveling the evolutionary history of Orangutans (genus: Pongo)- the impact of environmental processes and the genomic basis of adaptation Mattle-Greminger, Maja Patricia Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-121397 Dissertation Published Version Originally published at: Mattle-Greminger, Maja Patricia. Unraveling the evolutionary history of Orangutans (genus: Pongo)- the impact of environmental processes and the genomic basis of adaptation. 2015, University of Zurich, Faculty of Science. Unraveling the Evolutionary History of Orangutans (genus: Pongo) — The Impact of Environmental Processes and the Genomic Basis of Adaptation Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr. sc. nat.) vorgelegt der Mathematisch‐naturwissenschaftlichen Fakultät der Universität Zürich von Maja Patricia Mattle‐Greminger von Richterswil (ZH) Promotionskomitee Prof. Dr. Carel van Schaik (Vorsitz) PD Dr. Michael Krützen (Leitung der Dissertation) Dr. Maria Anisimova Zürich, 2015 To my family Table of Contents Table of Contents ........................................................................................................ 1 Summary ..................................................................................................................... 3 Zusammenfassung ..................................................................................................... -

Guidelines for Research and Data Collection Pan African Programme
Pan African Programme Data Collection Pan African Programme The cultured chimpanzee Guidelines for research and data collection This field protocol was a collaborative effort and greatly benefited from input from the following people: ARANDJELOVIC Mimi (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) BOESCH Christophe (Max Planck Institute for Evolutionary Anthropology / Wild Chimpanzee Foundation) CAMPBELL Geneviève (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) HOHMANN Gottfried (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) JUNKER Jessica (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) KOUAKOU Y. Célestin (Centre Suisse de Recherches Scientifiques en Côte-d’Ivoire / Wild Chimpanzee Foundation) KÜHL Hjalmar (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) LEENDERTZ Fabian (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) LEINERT Vera (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) MÖBIUS Yasmin (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) MURAI Mizuki (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) OELZE Victoria (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) RABANAL Louisa (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) ROBBINS Martha (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) VERGNES Virginie (Wild Chimpanzee Foundation) WAGNER Oliver (Max Planck Institute -
![Downloaded by [New York University] at 06:54 14 August 2016 Classic Case Studies in Psychology](https://docslib.b-cdn.net/cover/8368/downloaded-by-new-york-university-at-06-54-14-august-2016-classic-case-studies-in-psychology-738368.webp)
Downloaded by [New York University] at 06:54 14 August 2016 Classic Case Studies in Psychology
Downloaded by [New York University] at 06:54 14 August 2016 Classic Case Studies in Psychology The human mind is both extraordinary and compelling. But this is more than a collection of case studies; it is a selection of stories that illustrate some of the most extreme forms of human behaviour. From the leader who convinced his followers to kill themselves to the man who lost his memory; from the boy who was brought up as a girl to the woman with several personalities, Geoff Rolls illustrates some of the most fundamental tenets of psychology. Each case study has provided invaluable insights for scholars and researchers, and amazed the public at large. Several have been the inspiration for works of fiction, for example the story of Kim Peek, the real Rain Man. This new edition features three new case studies, including the story of Charles Decker who was tried for the attempted murder of two people but acquitted on the basis of a neurological condition, and Dorothy Martin, whose persisting belief in an impending alien invasion is an illuminating example of cognitive dissonance. In addition, each case study is contextualized with more typical behaviour, while the latest thinking in each sub-field is also discussed. Classic Case Studies in Psychology is accessibly written and requires no prior knowledge of psychology, but simply an interest in the human condition. It is a book that will amaze, sometimes disturb, but above all enlighten its readers. Downloaded by [New York University] at 06:54 14 August 2016 Geoff Rolls is Head of Psychology at Peter Symonds College in Winchester and formerly a Research Fellow at Southampton University, UK. -

West African Chimpanzees
Status Survey and Conservation Action Plan West African Chimpanzees Compiled and edited by Rebecca Kormos, Christophe Boesch, Mohamed I. Bakarr and Thomas M. Butynski IUCN/SSC Primate Specialist Group IUCN The World Conservation Union Donors to the SSC Conservation Communications Programme and West African Chimpanzees Action Plan The IUCN Species Survival Commission is committed to communicating important species conservation information to natural resource managers, decision makers and others whose actions affect the conservation of biodiversity. The SSC’s Action Plans, Occasional Papers, newsletter Species and other publications are supported by a wide variety of generous donors including: The Sultanate of Oman established the Peter Scott IUCN/SSC Action Plan Fund in 1990. The Fund supports Action Plan development and implementation. To date, more than 80 grants have been made from the Fund to SSC Specialist Groups. The SSC is grateful to the Sultanate of Oman for its confidence in and support for species conservation worldwide. The Council of Agriculture (COA), Taiwan has awarded major grants to the SSC’s Wildlife Trade Programme and Conser- vation Communications Programme. This support has enabled SSC to continue its valuable technical advisory service to the Parties to CITES as well as to the larger global conservation community. Among other responsibilities, the COA is in charge of matters concerning the designation and management of nature reserves, conservation of wildlife and their habitats, conser- vation of natural landscapes, coordination of law enforcement efforts, as well as promotion of conservation education, research, and international cooperation. The World Wide Fund for Nature (WWF) provides significant annual operating support to the SSC. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Intimate Relations—Mitochondria and Ageing
International Journal of Molecular Sciences Review Intimate Relations—Mitochondria and Ageing Michael Webb and Dionisia P. Sideris * Mitobridge Inc., an Astellas Company, 1030 Massachusetts Ave, Cambridge, MA 02138, USA; [email protected] * Correspondence: [email protected] Received: 29 August 2020; Accepted: 6 October 2020; Published: 14 October 2020 Abstract: Mitochondrial dysfunction is associated with ageing, but the detailed causal relationship between the two is still unclear. Wereview the major phenomenological manifestations of mitochondrial age-related dysfunction including biochemical, regulatory and energetic features. We conclude that the complexity of these processes and their inter-relationships are still not fully understood and at this point it seems unlikely that a single linear cause and effect relationship between any specific aspect of mitochondrial biology and ageing can be established in either direction. Keywords: mitochondria; ageing; energetics; ROS; gene regulation 1. Introduction The last two decades have witnessed a dramatic transformation in our view of mitochondria, their basic biology and functions. While still regarded as functioning primarily as the eukaryotic cell’s generator of energy in the form of adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide (reduced form; NADH), mitochondria are now recognized as having a plethora of functions, including control of apoptosis, regulation of calcium, forming a signaling hub and the synthesis of various bioactive molecules. Their biochemical functions beyond ATP supply include biosynthesis of lipids and amino acids, formation of iron sulphur complexes and some stages of haem biosynthesis and the urea cycle. They exist as a dynamic network of organelles that under normal circumstances undergo a constant series of fission and fusion events in which structural, functional and encoding (mtDNA) elements are subject to redistribution throughout the network. -
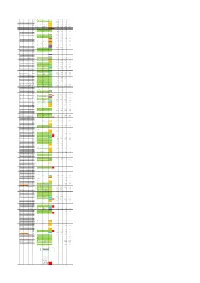
End Strand of Gene Gene Name Gene Function Starnd of Trascript
Genomic TSS strand Gene starnd of Transcipt relatively to TSS Start End Gene name coordinates relatively TSS type Control_fwd TEX_fwd Control_rev TEX_rev of gene function trascript gene group of TSS to ATG ribosomal 93 854 + rps12 + 1 protein S12 1 1 14.0638317 20.7539794 0 0 93 324 exon 1 + 111 rps12 intron 1 1 13.90756687 18.07149224 0.781323982 0.423550599 829 854 exon 2 + 496 rps12 exon2 2 24.22104343 15.24782157 0 0 30S 904 1371 + gene rps7 ribosomal + 2 protein S7 1303 -209 rps7 inter (ndhB) 2 20.47068832 9.035746118 0.625059185 0.847101199 NADH dehydrogen 1512 3535 + gene ndhB (ndh2) + ase subunit 3 2 1696 ndhB exon 1-inter 2 3.594090315 2.964854195 0.468794389 0.282367066 + 2209 ndhB exon 1-inter 2 46.09811492 4.09432246 0.468794389 0.423550599 1512 2237 exon 1 + 2756 ndhB exon 2 -inter 2 43.28534858 4.800240125 0.312529593 0.282367066 2756 3535 exon 2 + 3090 ndhB exon 2 -inter 2 17.50165719 15.95373924 0.312529593 0 + 3192 ndhB exon 2 -inter 2 140.6383167 117.6058831 2.812766334 1.694202397 - 3462 ndhB exon 2 -inter 3 1.406383167 1.129468265 1.406383167 3.67077186 4 3633 3712 + tRNA tRNA-Leu-CAA + 3610 -23 tRNA-Leu 1 77.19480938 84.85130339 0.625059185 0 + 3633 1 tRNA-Leu 1 359.5652963 649.0207016 0.781323982 0 photosyste 3954 4058 + gene psbM m II protein + 5 M 3775 -179 psbM 2 20.47068832 12.00060031 0 0.141183533 + 3954 psbM-0 2 69.22530477 28.37789015 0.156264796 0 hypothetical 4182 5073 + gene ycf66 6 protein 4182 4287 exon 1 4772 5073 exon 2 7 5202 5113 - gene ycf (ORF29) - 5299 orf29 inter 1 0 0 3.125295926 3.67077186 -

WO 2016/200987 Al 15 December 2016 (15.12.2016) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/200987 Al 15 December 2016 (15.12.2016) P O P C T (51) International Patent Classification: Trudi, A.; Indigo Agriculture, Inc., 500 Rutherford Aven A01N 63/02 (2006.01) A01C 1/00 (2006.01) ue, North Building, Boston, MA 02129 (US). A01N 63/00 (2006.01) A01C 1/06 (2006.01) (74) Agents: HUBL, Susan, T. et al; Fenwick & West LLP, (21) International Application Number: Silicon Valley Center, 801 California Street, Mountain PCT/US20 16/036504 View, CA 94041 (US). (22) International Filing Date: (81) Designated States (unless otherwise indicated, for every 8 June 2016 (08.06.2016) kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, (25) Filing Language: English BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, (26) Publication Language: English DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, N , IR, IS, JP, KE, KG, KN, KP, KR, (30) Priority Data: KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, 62/172,748 8 June 2015 (08.06.2015) US MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, 62/172,750 8 June 2015 (08.06.2015) US PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, 62/172,755 8 June 2015 (08.06.2015) us SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 62/3 16,386 31 March 2016 (3 1.03.2016) us TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Transcriptomic Regulation of Alternative Phenotypic Trajectories in Embryos of the Annual Killifish Austrofundulus Limnaeus
Portland State University PDXScholar Dissertations and Theses Dissertations and Theses Fall 11-30-2017 Transcriptomic Regulation of Alternative Phenotypic Trajectories in Embryos of the Annual Killifish Austrofundulus limnaeus Amie L. Romney Portland State University Follow this and additional works at: https://pdxscholar.library.pdx.edu/open_access_etds Part of the Biology Commons, and the Genetics and Genomics Commons Let us know how access to this document benefits ou.y Recommended Citation Romney, Amie L., "Transcriptomic Regulation of Alternative Phenotypic Trajectories in Embryos of the Annual Killifish Austrofundulus limnaeus" (2017). Dissertations and Theses. Paper 4033. https://doi.org/10.15760/etd.5917 This Dissertation is brought to you for free and open access. It has been accepted for inclusion in Dissertations and Theses by an authorized administrator of PDXScholar. Please contact us if we can make this document more accessible: [email protected]. Transcriptomic Regulation of Alternative Phenotypic Trajectories in embryos of the Annual Killifish Austrofundulus limnaeus by Amie Lynn Thomas Romney A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biology Dissertation Committee Jason Podrabsky, Chair Suzanne Estes Bradley Buckley Todd Rosenstiel Dirk Iwata-Reuyl Portland State University 2017 © 2017 Amie Lynn Thomas Romney ABSTRACT The Annual Killifish, Austrofundulus limnaeus, survives the seasonal drying of their pond habitat in the form of embryos entering diapause midway through development. The diapause trajectory is one of two developmental phenotypes. Alternatively, individuals can “escape” entry into diapause and develop continuously until hatching. The alternative phenotypes of A. limnaeus are a form of developmental plasticity that provides this species with a physiological adaption for surviving stressful environments.