The Matrix Method of Representation, Analysis and Classification of Long
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterization of a Full-Length Infectious Clone of Bovine Foamy Virus 3026
VIROLOGICA SINICA 2014, 29 (2): 94-102 DOI 10.1007/s12250-014-3382-5 RESEARCH ARTICLE Characterization of a full-length infectious clone of bovine foamy virus 3026 * Tiejun Bing1, Hong Yu1, 2, Yue Li1, Lei Sun3, Juan Tan1, Yunqi Geng1, Wentao Qiao1 1. Key Laboratory of Molecular Microbiology and Biotechnology (Ministry of Education) and Key Laboratory of Microbial Functional Genomics (Tianjin), College of Life Sciences, Nankai University, Tianjin 300071, China; 2. Department of Molecular and Cellular Pharmacology, the Vascular Biology Institute, University of Miami School of Medicine, Miami, FL 33136, USA; 3. National Laboratory of Biomacromolecules, Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China The biological features of most foamy viruses (FVs) are poorly understood, including bovine foamy virus (BFV). BFV strain 3026 (BFV3026) was isolated from the peripheral blood mononuclear cells of an infected cow in Zhangjiakou, China. A full-length genomic clone of BFV3026 was obtained from BFV3026-infected cells, and it exhibited more than 99% amino acid (AA) homology to another BFV strain isolated in the USA. Upon transfection into fetal canine thymus cells, the full-length BFV3026 clone produced viral structural and auxiliary proteins, typical cytopathic effects, and virus particles. These results demonstrate that the full-length BFV3026 clone is fully infectious and can be used in further BFV3026 research. KEYWORDS bovine foamy virus; infectious clone; syncytium; electron microscopy pol, and env structural genes and additional open read- INTRODUCTION ing frames (ORFs) that are under the control of the 5′- long terminal repeat (LTR) and an internal promoter (IP) Foamy viruses (FVs) are members of Retroviridae. -

Eukaryote Cell Biology - Michelle Gehringer
FUNDAMENTALS OF BIOCHEMISTRY, CELL BIOLOGY AND BIOPHYSICS – Vol. II - Eukaryote Cell Biology - Michelle Gehringer EUKARYOTE CELL BIOLOGY Michelle Gehringer Department of Biochemistry and Microbiology, University of Port Elizabeth, South Africa Keywords: cell theory, cell diversity, eukaryote cell structure, nucleus, chromatin, DNA, organelles, mitochondria, chloroplasts, transcription, RNA, translation, ribosomes, cell cycle, interphase, mitosis, meiosis, signal transduction, growth regulation, cancer, oncogenesis. Contents 1. Introduction 1.1. The first cell 2. Origin of Eukaryotes 3. Cellular differentiation in multicellular organisms 3.1. Plants 3.2. Animals 4. Eukaryotic cell structure 5. Organization of eukaryotic cells 5.1. Plasma membrane 5.2. Extracellular matrices 5.3. Protein synthesis and transport 5.4. Cytoskeleton and movement 5.5. Nucleus 5.5.1 Genomes 5.5.2 Gene expression 5.5.3 Maintaining the genome 5.6. Organelles 6. The cell cycle 6.1. Mitosis 6.2. Meiosis 7. Regulation of cell growth 7.1. Signal transduction 7.2. Programmed cell death 7.3. CancerUNESCO – EOLSS 8. Experimental Models 8.1. Yeast SAMPLE CHAPTERS 8.2. Arabidopsis 8.3. Drosophila 8.4. The mouse 8.5. Cell culture 8.6. Separation of cellular contents 8.7. Tracing biochemical pathways 9. Future Investigations Glossary Bibliography ©Encyclopedia of Life Support Systems (EOLSS) FUNDAMENTALS OF BIOCHEMISTRY, CELL BIOLOGY AND BIOPHYSICS – Vol. II - Eukaryote Cell Biology - Michelle Gehringer Biographical Sketch Summary Cells form the basic unit of life on our planet. They are well organized systems which perform all the essential tasks of eating, respiring, replicating and excreting waste products. The first cells, which are thought to have evolved about 3.8 billion years ago, much resembled present day prokaryotes. -

Characterization of Biofilm Extracts from Two Marine Bacteria
applied sciences Article Characterization of Biofilm Extracts from Two Marine Bacteria Delphine Passerini 1, Florian Fécamp 1, Laetitia Marchand 1, Laetitia Kolypczuk 1 , Sandrine Bonnetot 1, Corinne Sinquin 1 ,Véronique Verrez-Bagnis 1 , Dominique Hervio-Heath 2 , Sylvia Colliec-Jouault 1 and Christine Delbarre-Ladrat 1,* 1 Ifremer, Atlantique Center, Microbial Ecosystems and Marine Molecules for the Biotechnologies, 44311 Rue de l’Ile d’Yeu, Nantes CEDEX 3, France; [email protected] (D.P.); fl[email protected] (F.F.); [email protected] (L.M.); [email protected] (L.K.); [email protected] (S.B.); [email protected] (C.S.); [email protected] (V.V.-B.); [email protected] (S.C.-J.) 2 Ifremer, Bretagne Center, Health, Environment and Microbiology, 1625 Route de Sainte-Anne, 29280 Plouzané, France; [email protected] * Correspondence: [email protected] Received: 17 September 2019; Accepted: 13 November 2019; Published: 19 November 2019 Featured Application: By analyzing extracts of biofilm formed by two marine bacteria and comparing them with planktonic extracts, we have shown that biofilm may induce the biosynthesis of potentially bioactive compounds and may open up new possibilities for compound discovery. Abstract: In the marine environment, biofilm formation is an important lifestyle for microorganisms. A biofilm is comprised of cells embedded in an extracellular matrix that holds them close together and keeps the biofilm attached to the colonized surface. This predominant lifestyle and its main regulation pathway, namely quorum-sensing (QS), have been shown to induce specific bioactive metabolites. In this study, we investigated the biofilm formation by two marine bacteria belonging to the Vibrio species to discover potentially innovative bioactive compounds. -

The Origin of the Eukaryotic Cell Based on Conservation of Existing
The Origin of the Eukaryotic Albert D. G. de Roos The Beagle Armada Cell Based on Conservation Bioinformatics Division of Existing Interfaces Einsteinstraat 67 3316GG Dordrecht, The Netherlands [email protected] Abstract Current theories about the origin of the eukaryotic Keywords cell all assume that during evolution a prokaryotic cell acquired a Evolution, nucleus, eukaryotes, self-assembly, cellular membranes nucleus. Here, it is shown that a scenario in which the nucleus acquired a plasma membrane is inherently less complex because existing interfaces remain intact during evolution. Using this scenario, the evolution to the first eukaryotic cell can be modeled in three steps, based on the self-assembly of cellular membranes by lipid-protein interactions. First, the inclusion of chromosomes in a nuclear membrane is mediated by interactions between laminar proteins and lipid vesicles. Second, the formation of a primitive endoplasmic reticulum, or exomembrane, is induced by the expression of intrinsic membrane proteins. Third, a plasma membrane is formed by fusion of exomembrane vesicles on the cytoskeletal protein scaffold. All three self-assembly processes occur both in vivo and in vitro. This new model provides a gradual Darwinistic evolutionary model of the origins of the eukaryotic cell and suggests an inherent ability of an ancestral, primitive genome to induce its own inclusion in a membrane. 1 Introduction The origin of eukaryotes is one of the major challenges in evolutionary cell biology. No inter- mediates between prokaryotes and eukaryotes have been found, and the steps leading to eukaryotic endomembranes and endoskeleton are poorly understood. There are basically two competing classes of hypotheses: the endosymbiotic and the autogenic. -

Sensitivity of Golgi Matrix Proteins to an ER Exit Block
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central JCBArticle Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block Suzanne Miles,1 Heather McManus,1 Kimberly E. Forsten,2 and Brian Storrie1 1Department of Biochemistry and 2Department of Chemical Engineering, Virginia Tech, Blacksburg, VA 24061 e tested whether the entire Golgi apparatus is concentration. Redistribution of GalNAcT2 was more a dynamic structure in interphase mammalian sensitive to low Sar1pdn concentrations than giantin or Wcells by assessing the response of 12 different GM130. Redistribution was most rapid for p27, COPI, and Golgi region proteins to an endoplasmic reticulum (ER) exit p115. Giantin, GM130, and GalNAcT2 relocated with block. The proteins chosen spanned the Golgi apparatus approximately equal kinetics. Distinct ER accumulation and included both Golgi glycosyltransferases and putative could be demonstrated for all integral membrane proteins. matrix proteins. Protein exit from ER was blocked either by ER-accumulated Golgi region proteins were functional. microinjection of a GTP-restricted Sar1p mutant protein in Photobleaching experiments indicated that Golgi-to-ER the presence of a protein synthesis inhibitor, or by plasmid- protein cycling occurred in the absence of any ER exit encoded expression of the same dominant negative Sar1p. block. We conclude that the entire Golgi apparatus is a All Golgi region proteins examined lost juxtanuclear Golgi dynamic structure and suggest that most, if not all, Golgi apparatus–like distribution as scored by conventional and region–integral membrane proteins cycle through ER in confocal fluorescence microscopy in response to an ER interphase cells. -

Center of Biomedical Research Excellence in Matrix Biology: Building Research Infrastructure, Supporting Young Researchers, and Fostering Collaboration
International Journal of Molecular Sciences Editorial Center of Biomedical Research Excellence in Matrix Biology: Building Research Infrastructure, Supporting Young Researchers, and Fostering Collaboration Julia Thom Oxford 1,2,3,* , Ken A. Cornell 1,3,4, Jared J. Romero 1,5 , Diane B. Smith 1,3 , Tracy L. Yarnell 1,3, Rhiannon M. Wood 1,3 , Cheryl L. Jorcyk 1,2, Trevor J. Lujan 1,6, Allan R. Albig 1,2 , Kristen A. Mitchell 1,2, Owen M. McDougal 1,4 , Daniel Fologea 1,7, David Estrada 1,8, Juliette K. Tinker 1,2, Rajesh Nagarajan 1,4, Don L. Warner 1,4, Troy T. Rohn 1,2, Jim Browning 1,9 , Richard S. Beard Jr. 1,3, Lisa R. Warner 1,4 , Brad E. Morrison 1,2, Clare K. Fitzpatrick 1,6, Gunes Uzer 1,6, Laura Bond 1,3, Stephanie M. Frahs 1,3, Cynthia Keller-Peck 1,3, Xinzhu Pu 1,3, Luke G. Woodbury 1,3 and Matthew W. Turner 1,3 1 Center of Biomedical Research Excellence in Matrix Biology, Boise State University, Boise, ID 83725, USA 2 Department of Biological Sciences, Boise State University, Boise, ID 83725, USA 3 Biomolecular Research Center, Boise State University, Boise, ID 83725, USA 4 Department of Chemistry and Biochemistry, Boise State University, Boise, ID 83725, USA 5 Division of Research, Boise State University, Boise, ID 83725, USA 6 Department of Mechanical and Biomedical Engineering, Boise State University, Boise, ID 83725, USA 7 Department of Physics, Boise State University, Boise, ID 83725, USA 8 Micron School of Materials Science and Engineering, Boise State University, Boise, ID 83725, USA 9 Department of Electrical and Computer Engineering, Boise State University, Boise, ID 83725, USA * Correspondence: [email protected]; Tel.: +01-208-426-2395 Received: 9 March 2020; Accepted: 17 March 2020; Published: 20 March 2020 Abstract: The Center of Biomedical Research Excellence in Matrix Biology strives to improve our understanding of extracellular matrix at molecular, cellular, tissue, and organismal levels to generate new knowledge about pathophysiology, normal development, and regenerative medicine. -
Basic Features of All Cells
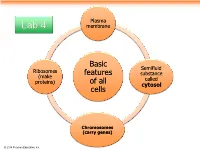
Plasma Lab 4 membrane Basic Semifluid Ribosomes features substance (make called proteins) of all cytosol cells Chromosomes (carry genes) © 2014 Pearson Education, Inc. Prokaryotic cells are characterized by having . No nucleus . DNA in an unbound region called the nucleoid . No membrane-bound organelles . Cytoplasm bound by the plasma membrane © 2014 Pearson Education, Inc. Eukaryotic cells are characterized by having • DNA in a nucleus that is bounded by a membranous nuclear envelope • Membrane-bound organelles • Cytoplasm in the region between the plasma membrane and nucleus Eukaryotic cells are generally much larger than prokaryotic cells © 2014 Pearson Education, Inc. Figure 6.8a ENDOPLASMIC RETICULUM (ER) Nuclear envelope Rough ER Smooth ER Nucleolus NUCLEUS Flagellum Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Lysosome Mitochondrion © 2014 Pearson Education, Inc. Figure 6.8b Nuclear envelope NUCLEUS Nucleolus Rough ER Chromatin Smooth ER Ribosomes Golgi Central vacuole apparatus Microfilaments CYTOSKELETON Microtubules Mitochondrion Peroxisome Plasma Chloroplast membrane Cell wall Plasmodesmata Wall of adjacent cell © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. Table 6.1 © 2014 Pearson Education, Inc. Cell Walls of Plants The cell wall is an extracellular structure that distinguishes plant cells from animal cells Plant cell walls Prokaryotes, are made -

An Endogenous Foamy-Like Viral Element in the Coelacanth Genome
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central An Endogenous Foamy-like Viral Element in the Coelacanth Genome Guan-Zhu Han*, Michael Worobey* Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, Arizona, United States of America Abstract Little is known about the origin and long-term evolutionary mode of retroviruses. Retroviruses can integrate into their hosts’ genomes, providing a molecular fossil record for studying their deep history. Here we report the discovery of an endogenous foamy virus-like element, which we designate ‘coelacanth endogenous foamy-like virus’ (CoeEFV), within the genome of the coelacanth (Latimeria chalumnae). Phylogenetic analyses place CoeEFV basal to all known foamy viruses, strongly suggesting an ancient ocean origin of this major retroviral lineage, which had previously been known to infect only land mammals. The discovery of CoeEFV reveals the presence of foamy-like viruses in species outside the Mammalia. We show that foamy-like viruses have likely codiverged with their vertebrate hosts for more than 407 million years and underwent an evolutionary transition from water to land with their vertebrate hosts. These findings suggest an ancient marine origin of retroviruses and have important implications in understanding foamy virus biology. Citation: Han G-Z, Worobey M (2012) An Endogenous Foamy-like Viral Element in the Coelacanth Genome. PLoS Pathog 8(6): e1002790. doi:10.1371/ journal.ppat.1002790 Editor: Michael Emerman, Fred Hutchinson Cancer Research Center, United States of America Received February 29, 2012; Accepted May 22, 2012; Published June 28, 2012 Copyright: ß 2012 Han, Worobey. -

Characterization of Giardia Cell Nucleus: Its Implication on the Nature and Origin of the Primitive Cell Nucleus
Cell Research(1995),5,115—124 Characterization of Giardia cell nucleus: Its implication on the nature and origin of the primitive cell nucleus LI JINGYAN Department of Evolutionary Cell Biology, Laboratory of Cellular and Molecular Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China. Key words: Origin of cell nucleus, nucleus of Giardia, primitive cell nu- cleus, archezoa. The central problem of the origin and early evolution of the eukaryotic cell is the origin of the cell nucleus, because the cell nucleus, with its double-layered nuclear envelope, is the most prominent and important morphological mark distinguishing eukaryotic cells from prokaryotes. Eukaryotic cells may have no chloroplast, mito- chondria or flagellum, but they must possess a cell nucleus. The cell nucleus itself is a very complicated structure. If we try to make clear the origin and early evolution of the cell nucleus, we ought to investigate the origin and evolution of each morphological component of a cell nucleus. My monograph on‘The genesis of eukaryotic cells in the evolutionary history of life’(1979, Science Press, Beijing, in Chinese)[1], mainly attempted to make some effort in this regard. However, to date, the hypothetical model of a fully depicted nucleus of a primitive cell has not been constructed. It seems likely that our recent studies of the nucleus of Giardia (diplomonad) have provided a realistic basis to construct such a model. I. The characteristics of the nucleus of Giardia Kingdom Archezoa (Cavalier-Smith, 1989), contains those parasitic and free- living protists which originally possess no mitochondria and no typical Golgi ap- paratus, but they have a 70s type of ribosomes containing 16s and 23s rRNAs, just as prokaryotes have. -

Extracellular Matrix Plays an Important Role in Stem Cell Biology
ell Res C ea m rc te h S & f Wen et al., J Stem Cell Res Ther 2018, 8:5 o T h l Journal of e a r n DOI: 10.4172/2157-7633.1000e122 a r p u y o J ISSN: 2157-7633 Stem Cell Research & Therapy Editorial Open Access Extracellular Matrix Plays an Important Role in Stem Cell Biology Yi Wen1,2, Sijun Hu3, Junjie Wu1,2* 1State Key Laboratory of Military Stomatology, Xi'an, Shaanxi Province, People's Republic of China 2Department of Orthodontics, Fourth Military Medical University, School of Stomatology, Xi'an, Shaanxi Province, People's Republic of China 3State Key Laboratory of Cancer Biology and Xijing Hospital of Digestive Diseases, The Fourth Military Medical University, Xi'an, Shaanxi Province, China Keywords: Extracellular matrix; Stem cell biology; Stem cells; Tissue as well as stem cell differentiation. Thus, ECM affects cell processes, and even gene expression through different stimulus [19]. In addition, it is Editorial worth mentioning that, ECM is also involved in cell death process. In In vivo stem cells live in a complex microenvironment called stem fact, programmed cell death, also known as anoikis, is due to a decrease in the interaction between the cells and ECM which is resulting from cell niche [1-3]. Stem cell niche can affect stem cell behavior and regulate stem cell fate by multiple signals. These signals may be structural, detachment between cells and ECM [20]. physical, electrical or biochemical. In the human body, the stem cell It is demonstrated by Kevin Lynch and Ming Pei [21] that the microenvironment can maintain adult stem cell in a quiescent state. -

Chondrogenesis, Studied with the Electron Microscope
CHONDROGENESIS, STUDIED WITH THE ELECTRON MICROSCOPE GABRIEL C. GODMAN, M.1)., and KEITH R. PORTER, Ph.I). From The Rockefeller Institute. Dr. Godman's present address is Department of Microbiology, Columbia University, New York ABSTRACT The role of the cells in the fabrication of a connective tissue matrix, and the structural modifications which accompany cytodifferentiation have been investigated in developing epiphyseal cartilage of fetal rat by means of electron microscopy. Differentiation of the pre- chondral mesenchymal cells to chondroblasts is marked by the acquisition of an extensive endoplasmic reticulum, enlargement and concentration of the Golgi apparatus, the ap- pearance of membrane-bounded cytoplasmic inclusions, and the formation of specialized foci of increased density in the cell cortex. These modifications are related to the secretion of the cartilage matrix. The matrix of young hyaline cartilage consists of groups of rela- tively short, straight, banded collagen fibrils of 10 to 20 m# and a dense granular component embedded in an amorphous ground substance of moderate electron density. It is postu- lated that the first phase of fibrillogenesis takes place at the cell cortex in dense bands or striae within the ectoplasm subjacent to the cell membrane. These can be resolved into sheaves of "primary" fibrils of about 7 to 10 m#. They are supposedly shed (by excortica- tion) into the matrix space between the separating chondroblasts, where they may serve as "cores" of the definitive matrix fibrils. The diameter of the fibrils may subsequently increase up to threefold, presumably by incorporation of "soluble" or tropocollagen units from the ground substance. The chondroblast also discharges into the matrix the electron- dense amorphous or granular contents of vesicles derived from the Golgi apparatus, and the mixed contents of large vacuoles or blebs bounded by distinctive double membranes. -

Between Retroviruses and Pararetroviruses
Virology 271, 1–8 (2000) doi:10.1006/viro.2000.0216, available online at http://www.idealibrary.com on MINIREVIEW Foamy Viruses: Between Retroviruses and Pararetroviruses Charles-Henri Lecellier and Ali Saı¨b 1 CNRS UPR9051, Universite´Paris 7, Hoˆpital Saint-Louis, 75475 Paris Cedex 10, France Received November 18, 1999; returned to author for revision December 14, 1999; accepted January 21, 2000 Foamy viruses, also called spumaviruses or spuma- sory proteins from the 3Ј end of the viral genome (Reth- retroviruses, are retroviruses described for the first time wilm et al., 1987). Construction of infectious clones of in 1954 in cell cultures derived from monkey kidneys HFV and cloning of viral genes out of the viral context (Enders et al., 1954). They are so named because of the allowed the study of the viral replication cycle and the specific cytopathic foam effect they induce in culture, implication of each gene product. One remarkable find- with the concomitant appearance of syncytia, facilitating ing is the striking similarity between FVs and the hepa- their isolation. In culture, FVs are highly lytic and present titis B virus (HBV, the prototype of hepadnaviruses) con- a large cellular tropism that reflects the ubiquity of the cerning the strategy used to replicate their genome (See- viral receptor (Hill et al., 1999). These viruses were ger et al., 1996). mainly isolated from primate species and also from non- The Third International Conference on Foamy Viruses, primates, such as cats (feline foamy virus [FeFV]) or held in Paris in June 1999, has brought new insights into cattle (bovine foamy virus [BFV]) and more recently from our understanding of these particular viruses, confirming horses (Tobaly-Tapiero et al., 1999) (Table 1).