Supplemental Material
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Imm Catalog.Pdf
$ Gene Symbol A B 3 C 4 D 9 E 10 F 11 G 12 H 13 I 14 J. K 17 L 18 M 19 N 20 O. P 22 R 26 S 27 T 30 U 32 V. W. X. Y. Z 33 A ® ® Gene Symbol Gene ID Antibody Monoclonal Antibody Polyclonal MaxPab Full-length Protein Partial-length Protein Antibody Pair KIt siRNA/Chimera Gene Symbol Gene ID Antibody Monoclonal Antibody Polyclonal MaxPab Full-length Protein Partial-length Protein Antibody Pair KIt siRNA/Chimera A1CF 29974 ● ● ADAMTS13 11093 ● ● ● ● ● A2M 2 ● ● ● ● ● ● ADAMTS20 80070 ● AACS 65985 ● ● ● ADAMTS5 11096 ● ● ● AANAT 15 ● ● ADAMTS8 11095 ● ● ● ● AATF 26574 ● ● ● ● ● ADAMTSL2 9719 ● AATK 9625 ● ● ● ● ADAMTSL4 54507 ● ● ABCA1 19 ● ● ● ● ● ADAR 103 ● ● ABCA5 23461 ● ● ADARB1 104 ● ● ● ● ABCA7 10347 ● ADARB2 105 ● ABCB9 23457 ● ● ● ● ● ADAT1 23536 ● ● ABCC4 10257 ● ● ● ● ADAT2 134637 ● ● ABCC5 10057 ● ● ● ● ● ADAT3 113179 ● ● ● ABCC8 6833 ● ● ● ● ADCY10 55811 ● ● ABCD2 225 ● ADD1 118 ● ● ● ● ● ● ABCD4 5826 ● ● ● ADD3 120 ● ● ● ABCG1 9619 ● ● ● ● ● ADH5 128 ● ● ● ● ● ● ABL1 25 ● ● ADIPOQ 9370 ● ● ● ● ● ABL2 27 ● ● ● ● ● ADK 132 ● ● ● ● ● ABO 28 ● ● ADM 133 ● ● ● ABP1 26 ● ● ● ● ● ADNP 23394 ● ● ● ● ABR 29 ● ● ● ● ● ADORA1 134 ● ● ACAA2 10449 ● ● ● ● ADORA2A 135 ● ● ● ● ● ● ● ACAN 176 ● ● ● ● ● ● ADORA2B 136 ● ● ACE 1636 ● ● ● ● ADRA1A 148 ● ● ● ● ACE2 59272 ● ● ADRA1B 147 ● ● ACER2 340485 ● ADRA2A 150 ● ● ACHE 43 ● ● ● ● ● ● ADRB1 153 ● ● ACIN1 22985 ● ● ● ADRB2 154 ● ● ● ● ● ACOX1 51 ● ● ● ● ● ADRB3 155 ● ● ● ● ACP5 54 ● ● ● ● ● ● ● ADRBK1 156 ● ● ● ● ACSF2 80221 ● ● ADRM1 11047 ● ● ● ● ACSF3 197322 ● ● AEBP1 165 ● ● ● ● ACSL4 2182 ● -

14321 NAE1/APPBP1 (D9I4Z) Rabbit Mab
Revision 1 C 0 2 - t NAE1/APPBP1 (D9I4Z) Rabbit mAb a e r o t S Orders: 877-616-CELL (2355) [email protected] 1 Support: 877-678-TECH (8324) 2 3 Web: [email protected] 4 www.cellsignal.com 1 # 3 Trask Lane Danvers Massachusetts 01923 USA For Research Use Only. Not For Use In Diagnostic Procedures. Applications: Reactivity: Sensitivity: MW (kDa): Source/Isotype: UniProt ID: Entrez-Gene Id: WB H M R Mk Endogenous 60 Rabbit IgG Q13564 8883 Product Usage Information 2. Huang, D.T. et al. (2005) Mol Cell 17, 341-50. 3. Liakopoulos, D. et al. (1998) EMBO J 17, 2208-14. Application Dilution 4. Gong, L. and Yeh, E.T. (1999) J Biol Chem 274, 12036-42. 5. Wada, H. et al. (2000) J Biol Chem 275, 17008-15. Western Blotting 1:1000 6. Sakata, E. et al. (2007) Nat Struct Mol Biol 14, 167-8. 7. Kawakami, T. et al. (2001) EMBO J 20, 4003-12. Storage 8. Podust, V.N. et al. (2000) Proc Natl Acad Sci USA 97, 4579-84. 9. Wu, K. et al. (2002) J Biol Chem 277, 516-27. Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA, 50% 10. Amir, R.E. et al. (2002) J Biol Chem 277, 23253-9. glycerol and less than 0.02% sodium azide. Store at –20°C. Do not aliquot the antibody. 11. Herrmann, J. et al. (2007) Circ Res 100, 1276-91. 12. Walden, H. et al. (2003) Mol Cell 12, 1427-37. -

Is APP Key to the Appbp1 Pathway?
Open Access Full Text Article Austin Alzheimer’s and Parkinson’s Disease A Austin Publishing Group Review Article Cycle on Wheels: Is APP Key to the AppBp1 Pathway? Chen Y1,2*, Neve RN4, Zheng H3, Griffin WST1,2, Barger SW1,2 and Mrak RE5 Abstract 1Department of Geriatrics, University of Arkansas for Alzheimer’s disease (AD) is the gradual loss of the cognitive function due Medical Sciences, USA to neuronal death. Currently no therapy is available to slow down, reverse 2Department of Neurobiology and Developmental or prevent the disease. Here we analyze the existing data in literature and Sciences, University of Arkansas for Medical Sciences, hypothesize that the physiological function of the Amyloid Precursor Protein USA (APP) is activating the AppBp1 pathway and this function is gradually lost during 3Huffington Center on Aging, Baylor College of Medicine, the progression of AD pathogenesis. The AppBp1 pathway, also known as the USA neddylation pathway, activates the small ubiquitin-like protein nedd8, which 4Department of Brain and Cognitive Sciences, covalently modifies and switches on Cullin ubiquitin ligases, which are essential Massachusetts Institute of Technology, USA in the turnover of cell cycle proteins. Here we discuss how APP may activate the 5Department of Pathology, University of Toledo Health AppBp1 pathway, which downregulates cell cycle markers and protects genome Sciences Campus, USA integrity. More investigation of this mechanism-driven hypothesis may provide *Corresponding author: Chen Y, Department insights into disease treatment and prevention strategies. of Geriatrics and Department of Neurobiology and Keywords: APP; Alzheimer’s disease; Ubiquitination; Neddylation; Cell Developmental Sciences, University of Arkansas for cycle Medical Sciences, Little Rock, AR 72205, USA Received: September 02, 2014; Accepted: September 29, 2014; Published: September 30, 2014 Abbreviations at least 18 proteins have been identified to bind AICD [25-27]. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Identification of SH3 Domain Proteins Interacting with the Cytoplasmic Tail of the a Disintegrin and Metalloprotease 10 (ADAM10)
Identification of SH3 Domain Proteins Interacting with the Cytoplasmic Tail of the A Disintegrin and Metalloprotease 10 (ADAM10) Henriette Ebsen, Marcus Lettau, Dieter Kabelitz, Ottmar Janssen* University of Kiel, Molecular Immunology, Institute for Immunology, University Hospital Schleswig-Holstein Campus Kiel, Kiel, Germany Abstract The a disintegrin and metalloproteases (ADAMs) play a pivotal role in the control of development, adhesion, migration, inflammation and cancer. Although numerous substrates of ADAM10 have been identified, the regulation of its surface expression and proteolytic activity is still poorly defined. One current hypothesis is that both processes are in part modulated by protein-protein interactions mediated by the intracellular portion of the protease. For related proteases, especially proline-rich regions serving as docking sites for Src homology domain 3 (SH3) domain-containing proteins proved to be important for mediating regulatory interactions. In order to identify ADAM10-binding SH3 domain proteins, we screened the All SH3 Domain Phager library comprising 305 human SH3 domains using a GST fusion protein with the intracellular region of human ADAM10 as a bait for selection. Of a total of 291 analyzed phage clones, we found 38 SH3 domains that were precipitated with the ADAM10-derived fusion protein but not with GST. We verified the binding to the cytosolic portion of ADAM10 for several candidates by co-immunoprecipitation and/or pull down analyses. Intriguingly, several of the identified proteins have been implicated in regulating surface appearance and/or proteolytic activity of related ADAMs. Thus, it seems likely that they also play a role in ADAM10 biology. Citation: Ebsen H, Lettau M, Kabelitz D, Janssen O (2014) Identification of SH3 Domain Proteins Interacting with the Cytoplasmic Tail of the A Disintegrin and Metalloprotease 10 (ADAM10). -
![APPBP1 (NAE1) Mouse Monoclonal Antibody [Clone ID: OTI1E10] Product Data](https://docslib.b-cdn.net/cover/0463/appbp1-nae1-mouse-monoclonal-antibody-clone-id-oti1e10-product-data-380463.webp)
APPBP1 (NAE1) Mouse Monoclonal Antibody [Clone ID: OTI1E10] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA804384 APPBP1 (NAE1) Mouse Monoclonal Antibody [Clone ID: OTI1E10] Product data: Product Type: Primary Antibodies Clone Name: OTI1E10 Applications: IHC, WB Recommended Dilution: WB 1:2000, IHC 1:150 Reactivity: Human, Mouse, Rat Host: Mouse Isotype: IgG1 Clonality: Monoclonal Immunogen: Human recombinant protein fragment corresponding to amino acids 1-274 of human NAE1 (NP_001018170) produced in E.coli. Formulation: PBS (PH 7.3) containing 1% BSA, 50% glycerol and 0.02% sodium azide. Concentration: 1 mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 50.4 kDa Gene Name: NEDD8 activating enzyme E1 subunit 1 Database Link: NP_001018170 Entrez Gene 84019 RatEntrez Gene 234664 MouseEntrez Gene 8883 Human Q13564 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 APPBP1 (NAE1) Mouse Monoclonal Antibody [Clone ID: OTI1E10] – TA804384 Background: The protein encoded by this gene binds to the beta-amyloid precursor protein. Beta-amyloid precursor protein is a cell surface protein with signal-transducing properties, and it is thought to play a role in the pathogenesis of Alzheimer's disease. In addition, the encoded protein can form a heterodimer with UBE1C and bind and activate NEDD8, a ubiquitin-like protein. -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

Genome-Wide DNA Methylation Analysis of KRAS Mutant Cell Lines Ben Yi Tew1,5, Joel K
www.nature.com/scientificreports OPEN Genome-wide DNA methylation analysis of KRAS mutant cell lines Ben Yi Tew1,5, Joel K. Durand2,5, Kirsten L. Bryant2, Tikvah K. Hayes2, Sen Peng3, Nhan L. Tran4, Gerald C. Gooden1, David N. Buckley1, Channing J. Der2, Albert S. Baldwin2 ✉ & Bodour Salhia1 ✉ Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 diferentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in diferentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream efector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer. Activating KRAS mutations can be found in nearly 25 percent of all cancers1. -
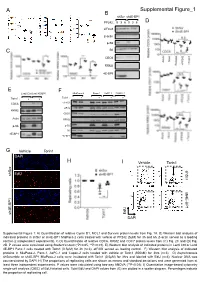
H Supplemental Figure 1 I D E B a C
A Supplemental Figure_1 B shScr sh4E-BP1 PP242 : 0 3 6 0 3 6 D eIF4G1 β-actin p-S6 C S6 CDC6 RRM2 4E-BP1 E F Lenti Ctrl Lenti 4E-BP1 MiaPaca-2 Panc-1 AsPC-1 Capan-2 Torin1 : Torin1 : + + + + + + CDC6 eIF4G1 eIF3a RRM2 CDC6 Actin RRM2 p-S6 p-S6 S6 4E-BP1 4E-BP1 G Vehicle Torin1 DAPI H I Vehicle Torin1 62.20% 45.30% shScr sh4E-BP1 EdU shScr 59.58% 49.47% EdU sh4E-BP1 DAPI Supplemental Figure 1: A) Quantification of relative Cyclin D1, MCL1 and Survivin protein levels from Fig. 1A. B) Western blot analysis of indicated proteins in shScr or sh4E-BP1 MiaPaca-2 cells treated with vehicle or PP242 (5µM) for 3h and 6h. β-actin served as a loading control (2 independent experiments). C-D) Quantification of relative CDC6, RRM2 and CDC7 protein levels from (C) Fig. 2C and (D) Fig. 2D. P values were calculated using Student’s t-test (*P<0.05, **P<0.01). E) Western blot analysis of indicated proteins in Lenti Ctrl or Lenti 4E-BP1 Panc-1 cells treated with Torin1 (0.5µM) for 3h (n=3). eIF4GI served as loading control. F) Western blot analysis of indicated proteins in MiaPaca-2, Panc-1, AsPC-1 and Capan-2 cells treated with vehicle or Torin1 (500nM) for 3hrs (n=3). G) Asynchronous shScramble or sh4E-BP1 MiaPaca-2 cells were incubated with Torin1 (0.5μM) for 3hrs and labeled with EdU (n=3). Nuclear DNA was counterstained by DAPI. H) The proportions of replicating cells are shown as means and standard deviations and were generated from at least three independent experiments. -

UNIVERSITY of PÉCS Characterization of the Vestibular NADPH
UNIVERSITY OF PÉCS Biological Doctoral School Characterization of the Vestibular NADPH Oxidase Enzyme Complex PhD Thesis PÉTER KISS PÉCS, 2009 1 UNIVERSITY OF PÉCS Biological Doctoral School Characterization of the Vestibular NADPH Oxidase Enzyme Complex PhD Thesis PÉTER KISS Supervisors : Dr. Joseph Zabner M.D. Dr. Kerepesi Ildikó, PhD PÉCS, 2009 2 1. Table of Contents 1. Table of Contents.....................................................................................3 2. Glossary and Abbreviations .....................................................................5 3. Introduction ..............................................................................................7 3.1 Reactive oxygen species......................................................................7 3.1.1 Reactive oxygen species in biology ...............................................7 3.1.2 Reactive oxygen species produced by NADPH oxidases..............9 3.1.2.1 Superoxide ..............................................................................9 3.1.2.2 Hydrogen peroxide................................................................10 3.1.2 The source of free radicals ..........................................................12 3.1.2.1 Endogenous free radicals......................................................13 3.1.2.1.1 Mitochondrial sources of free radicals ............................13 3.1.2.2 Extramitochondrial ROS sources ..........................................13 3.1.2.2.1 Cytochrome P-450 enzymes ..........................................13 -

Anti-CDK14 / PFTK1 Antibody (ARG40789)
Product datasheet [email protected] ARG40789 Package: 100 μl anti-CDK14 / PFTK1 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes CDK14 / PFTK1 Tested Reactivity Hu Tested Application FACS, IHC-P, WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name CDK14 / PFTK1 Antigen Species Human Immunogen KLH-conjugated synthetic peptide corresponding to aa. 67-96 of Human CDK14 / PFTK1. Conjugation Un-conjugated Alternate Names PFTAIRE1; Cell division protein kinase 14; hPFTAIRE1; EC 2.7.11.22; Cyclin-dependent kinase 14; PFTK1; Serine/threonine-protein kinase PFTAIRE-1 Application Instructions Application table Application Dilution FACS 1:10 - 1:50 IHC-P 1:50 - 1:100 WB 1:1000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 53 kDa Properties Form Liquid Purification Saturated Ammonium Sulfate (SAS) precipitation followed by dialysis against PBS. Buffer PBS and 0.09% (W/V) Sodium azide. Preservative 0.09% (W/V) Sodium azide. Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. www.arigobio.com 1/3 Bioinformation Gene Symbol CDK14 Gene Full Name cyclin-dependent kinase 14 Background PFTK1 is a member of the CDC2 (MIM 116940)-related protein kinase family (Yang and Chen, 2001 [PubMed 11313143]).[supplied by OMIM, Mar 2008] Function Serine/threonine-protein kinase involved in the control of the eukaryotic cell cycle, whose activity is controlled by an associated cyclin. -

NAE1 (APPBP1) Antibody (C-Term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap7273b
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 NAE1 (APPBP1) Antibody (C-term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP7273b Specification NAE1 (APPBP1) Antibody (C-term) - Product Information Application WB, IHC-P,E Primary Accession Q13564 Other Accession Q9Z1A5, Q8VBW6, Q4R3L6, NP_003896 Reactivity Human Predicted Monkey, Mouse, Rat Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Calculated MW 60246 Antigen Region 430-459 NAE1 (APPBP1) Antibody (C-term) - Additional Western blot analysis of anti-APPBP1 Information Antibody (C-term) (Cat.#AP7273b) in mouse brain tissue lysates (35ug/lane). Gene ID 8883 APPBP1(arrow) was detected using the purified Pab. Other Names NEDD8-activating enzyme E1 regulatory subunit, Amyloid beta precursor protein-binding protein 1, 59 kDa, APP-BP1, Amyloid protein-binding protein 1, Proto-oncogene protein 1, NAE1, APPBP1 Target/Specificity This NAE1 (APPBP1) antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 430-459 amino acids from the C-terminal region of human NAE1 (APPBP1). Dilution WB~~1:1000 IHC-P~~1:50~100 Western blot analysis of APP-BP1(arrow) using rabbit polyclonal APP-BP1 Antibody Format (C-term) (Cat.#AP7273b). 293 cell lysates (2 Purified monoclonal antibody supplied in ug/lane) either nontransfected (Lane 1) or PBS with 0.09% (W/V) sodium azide. This transiently transfected with the APP-BP1 antibody is purified through a protein G gene (Lane 2) (Origene Technologies). column, followed by dialysis against PBS. Storage Maintain refrigerated at 2-8°C for up to 2 Page 1/2 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 weeks.