Dasycercus Cristicauda)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Platypus Collins, L.R
AUSTRALIAN MAMMALS BIOLOGY AND CAPTIVE MANAGEMENT Stephen Jackson © CSIRO 2003 All rights reserved. Except under the conditions described in the Australian Copyright Act 1968 and subsequent amendments, no part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic, mechanical, photocopying, recording, duplicating or otherwise, without the prior permission of the copyright owner. Contact CSIRO PUBLISHING for all permission requests. National Library of Australia Cataloguing-in-Publication entry Jackson, Stephen M. Australian mammals: Biology and captive management Bibliography. ISBN 0 643 06635 7. 1. Mammals – Australia. 2. Captive mammals. I. Title. 599.0994 Available from CSIRO PUBLISHING 150 Oxford Street (PO Box 1139) Collingwood VIC 3066 Australia Telephone: +61 3 9662 7666 Local call: 1300 788 000 (Australia only) Fax: +61 3 9662 7555 Email: [email protected] Web site: www.publish.csiro.au Cover photos courtesy Stephen Jackson, Esther Beaton and Nick Alexander Set in Minion and Optima Cover and text design by James Kelly Typeset by Desktop Concepts Pty Ltd Printed in Australia by Ligare REFERENCES reserved. Chapter 1 – Platypus Collins, L.R. (1973) Monotremes and Marsupials: A Reference for Zoological Institutions. Smithsonian Institution Press, rights Austin, M.A. (1997) A Practical Guide to the Successful Washington. All Handrearing of Tasmanian Marsupials. Regal Publications, Collins, G.H., Whittington, R.J. & Canfield, P.J. (1986) Melbourne. Theileria ornithorhynchi Mackerras, 1959 in the platypus, 2003. Beaven, M. (1997) Hand rearing of a juvenile platypus. Ornithorhynchus anatinus (Shaw). Journal of Wildlife Proceedings of the ASZK/ARAZPA Conference. 16–20 March. -

Spotted Tailed Quoll (Dasyurus Maculatus)
Husbandry Guidelines for the SPOTTED-TAILED QUOLL (Tiger Quoll) (Photo: J. Marten) Dasyurus maculatus (MAMMALIA: DASYURIDAE) Author: Julie Marten Date of Preparation: February 2013 – June 2014 Western Sydney Institute of TAFE, Richmond Course Name and Number: Captive Animals Certificate III (18913) Lecturers: Graeme Phipps, Jacki Salkeld, Brad Walker DISCLAIMER Please note that this information is just a guide. It is not a definitive set of rules on how the care of Spotted- Tailed Quolls must be conducted. Information provided may vary for: • Individual Spotted-Tailed Quolls • Spotted-Tailed Quolls from different regions of Australia • Spotted-Tailed Quolls kept in zoos versus Spotted-Tailed Quolls from the wild • Spotted-Tailed Quolls kept in different zoos Additionally different zoos have their own set of rules and guidelines on how to provide husbandry for their Spotted-Tailed Quolls. Even though I researched from many sources and consulted various people, there are zoos and individual keepers, researchers etc. that have more knowledge than myself and additional research should always be conducted before partaking any new activity. Legislations are regularly changing and therefore it is recommended to research policies set out by national and state government and associations such as ARAZPA, ZAA etc. Any incident resulting from the misuse of this document will not be recognised as the responsibility of the author. Please use at the participants discretion. Any enhancements to this document to increase animal care standards and husbandry techniques are appreciated. Otherwise I hope this manual provides some helpful information. Julie Marten Picture J.Marten 2 OCCUPATIONAL HEALTH AND SAFETY RISKS It is important before conducting any work that all hazards are identified. -

SPOTTED-TAILED QUOLL (Tiger Quoll)
Husbandry Guidelines for the SPOTTED-TAILED QUOLL (Tiger Quoll) (Photo: J. Marten) Dasyurus maculatus (MAMMALIA: DASYURIDAE) Date By From Version 2014 Julie Marten WSI Richmond v 1 DISCLAIMER Please note that this information is just a guide. It is not a definitive set of rules on how the care of Spotted- Tailed Quolls must be conducted. Information provided may vary for: Individual Spotted-Tailed Quolls Spotted-Tailed Quolls from different regions of Australia Spotted-Tailed Quolls kept in zoos versus Spotted-Tailed Quolls from the wild Spotted-Tailed Quolls kept in different zoos Additionally different zoos have their own set of rules and guidelines on how to provide husbandry for their Spotted-Tailed Quolls. Even though I researched from many sources and consulted various people, there are zoos and individual keepers, researchers etc. that have more knowledge than myself and additional research should always be conducted before partaking any new activity. Legislations are regularly changing and therefore it is recommended to research policies set out by national and state government and associations such as ARAZPA, ZAA etc. Any incident resulting from the misuse of this document will not be recognised as the responsibility of the author. Please use at the participants discretion. Any enhancements to this document to increase anima l care standards and husbandry techniques are appreciated. Otherwise I hope this manual provides some helpful information. Julie Marten Picture J.Marten 2 OCCUPATIONAL HEALTH AND SAFETY RISKS It is important before conducting any work that all hazards are identified. This includes working with the animal and maintaining the enclosure. -

Phylogenetic Relationships of Living and Recently Extinct Bandicoots Based on Nuclear and Mitochondrial DNA Sequences ⇑ M
Molecular Phylogenetics and Evolution 62 (2012) 97–108 Contents lists available at SciVerse ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Phylogenetic relationships of living and recently extinct bandicoots based on nuclear and mitochondrial DNA sequences ⇑ M. Westerman a, , B.P. Kear a,b, K. Aplin c, R.W. Meredith d, C. Emerling d, M.S. Springer d a Genetics Department, LaTrobe University, Bundoora, Victoria 3086, Australia b Palaeobiology Programme, Department of Earth Sciences, Uppsala University, Villavägen 16, SE-752 36 Uppsala, Sweden c Australian National Wildlife Collection, CSIRO Sustainable Ecosystems, Canberra, ACT 2601, Australia d Department of Biology, University of California, Riverside, CA 92521, USA article info abstract Article history: Bandicoots (Peramelemorphia) are a major order of australidelphian marsupials, which despite a fossil Received 4 November 2010 record spanning at least the past 25 million years and a pandemic Australasian range, remain poorly Revised 6 September 2011 understood in terms of their evolutionary relationships. Many living peramelemorphians are critically Accepted 12 September 2011 endangered, making this group an important focus for biological and conservation research. To establish Available online 11 November 2011 a phylogenetic framework for the group, we compiled a concatenated alignment of nuclear and mito- chondrial DNA sequences, comprising representatives of most living and recently extinct species. Our Keywords: analysis confirmed the currently recognised deep split between Macrotis (Thylacomyidae), Chaeropus Marsupial (Chaeropodidae) and all other living bandicoots (Peramelidae). The mainly New Guinean rainforest per- Bandicoot Peramelemorphia amelids were returned as the sister clade of Australian dry-country species. The wholly New Guinean Per- Phylogeny oryctinae was sister to Echymiperinae. -

Wildlife Matters Wildlife Conservancy
australian wildlife matters wildlife conservancy Spring 2009 Pungalina reveals one of Australia’s rarest mammals Carpentarian Pseudantechinus 2 australian saving australia’s threatened wildlife wildlife Pictograph conservancy Welcome to the Spring 2009 edition of Wildlife Matters. As this edition goes to print, we are in the process of fi nalising the acquisition of Bowra (see pages 4-5), a 14,000 the awc mission hectare property located in the heart of the Mulga Lands in Queensland. Bowra will The mission of Australian Wildlife Conservancy be our 21st sanctuary, bringing the AWC network to more than 2.56 million hectares (AWC) is the effective conservation of all (6.3 million acres). Australian animal species and the habitats in While the overall scale of the portfolio is impressive, it is not the number of properties or which they live. To achieve this mission, our hectares that really count. A more accurate measure of the value of the portfolio is the actions are focused on: number of species and ecosystems that occur within the AWC estate. In this respect, • Establishing a network of sanctuaries the statistics are even more impressive – for example, around 80% of all Australian which protect threatened wildlife and terrestrial bird species and over 60% of all terrestrial mammal species occur on one or ecosystems: AWC now manages 20 more of our sanctuaries. sanctuaries covering over 2.56 million The fact that our portfolio captures such a high percentage of Australia’s wildlife species hectares (6.3 million acres). refl ects a deliberate, science-based strategy to ensure that AWC invests in properties • Implementing practical, on-ground of the highest environmental value. -

Terrestrial Native Mammals of Western Australia
TERRESTRIALNATIVE MAMMALS OF WESTERNAUSTRALIA On a number of occasionswe have been asked what D as y ce r cus u ist ica ud q-Mul Aara are the marsupialsof W.A. or what is the scientiflcname Anlechinusfla.t,ipes Matdo given to a palticular animal whosecommon name only A n t ec h i nus ap i ca I i s-Dlbbler rs known. Antechinusr osemondae-Little Red Antechinus As a guide,the following list of62 speciesof marsupials A nteclt itus mqcdonneIlens is-Red-eared Antechi nus and 59 speciesof othersis publishedbelow. Antechinus ? b ilar n i-Halney' s Antechinus Antec h in us mqculatrJ-Pismv Antechinus N ingaui r idei-Ride's Nirfaui - MARSUPALIA Ningauirinealvi Ealev's-KimNinsaui Ptaiigole*fuilissima beiiey Planigale Macropodidae Plani gale tenuirostris-Narrow-nosed Planigate Megaleia rufa Red Kangaroo Smi nt hopsis mu rina-Common Dulnart Macropus robustus-Etro Smin t hop[is longicaudat.t-Long-tailed Dunnart M acr opus fu Ii g inos,s-Western Grey Kangaroo Sminthops is cras sicaudat a-F at-tailed Dunnart Macrcpus antilo nus Antilope Kangaroo S-nint hopsi s froggal//- Larapinla Macropu"^agi /rs Sandy Wallaby Stnintllopsirgranuli,oer -Whire-railed Dunnart Macrcpus rirra Brush Wallaby Sninthopsis hir t ipes-Hairy -footed Dunnart M acro ptrs eugenii-T ammar Sminthopsiso oldea-^f r oughton's Dunnart Set oni x brac ltyuru s-Quokka A ntec h inomys lanrger-Wuhl-Wuhl On y ch oga I ea Lng uife r a-Kar r abul M.yr nte c o b ius fasc ialrls-N umbat Ony c hogalea Iunq ta-W \rrur.g Notoryctidae Lagorchest es conspic i Ilat us,Spectacied Hare-Wallaby Notorlctes -

Mulgara Husbandry Ma
Husbandry Guidelines Mulgara (Dasycercus cristicauda) Alice Springs Desert Park 2007 Compiled by Wes Caton CONTENTS: TAXONOMY.....................................................................................................................1 Common Name................................................................................................1.1 Classifacation...................................................................................................1.2 A.S.M.P. Category...........................................................................................1.3 I.U.C.N. Category............................................................................................1.4 O.H.&S. Category...........................................................................................1.5 Studbook Keeper.............................................................................................1.6 NATURAL HISTORY......................................................................................................2 Family..............................................................................................................2.1 DISTRIBUTION...............................................................................................................3 Map...................................................................................................................3.1 Habitat .............................................................................................................3.2 MORPHOMETRICS........................................................................................................4 -

Bandicoot Fossils and DNA Elucidate Lineage Antiquity Amongst Xeric
www.nature.com/scientificreports OPEN Bandicoot fossils and DNA elucidate lineage antiquity amongst xeric-adapted Received: 31 May 2016 Accepted: 31 October 2016 Australasian marsupials Published: 24 November 2016 Benjamin P. Kear1,2, Ken P. Aplin3 & Michael Westerman4 Bandicoots (Peramelemorphia) are a unique order of Australasian marsupials whose sparse fossil record has been used as prima facie evidence for climate change coincident faunal turnover. In particular, the hypothesized replacement of ancient rainforest-dwelling extinct lineages by antecedents of xeric-tolerant extant taxa during the late Miocene (~10 Ma) has been advocated as a broader pattern evident amongst other marsupial clades. Problematically, however, this is in persistent conflict with DNA phylogenies. We therefore determine the pattern and timing of bandicoot evolution using the first combined morphological + DNA sequence dataset of Peramelemorphia. In addition, we document a remarkably archaic new fossil peramelemorphian taxon that inhabited a latest Quaternary mosaic savannah-riparian forest ecosystem on the Aru Islands of Eastern Indonesia. Our phylogenetic analyses reveal that unsuspected dental homoplasy and the detrimental effects of missing data collectively obscure stem bandicoot relationships. Nevertheless, recalibrated molecular clocks and multiple ancestral area optimizations unanimously infer an early diversification of modern xeric-adapted forms. These probably originated during the late Palaeogene (30–40 Ma) alongside progenitors of other desert marsupials, and thus occupied seasonally dry heterogenous habitats long before the onset of late Neogene aridity. Bandicoots (Peramelemorphia) are a speciose order of Australasian marsupials that appeared early in the evolu- tionary history of Australidelphia1. Most are small to medium sized (up to 5 kg) terrestrial omnivores occupying a spectrum of rainforest to desert habitats2,3. -

Ecology and Predator Associations of the Northern Quoll (Dasyurus Hallucatus) in the Pilbara Lorna Hernandez Santin M.Sc
Ecology and predator associations of the northern quoll (Dasyurus hallucatus) in the Pilbara Lorna Hernandez Santin M.Sc. A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2016 School of Biological Sciences Abstract The northern quoll (Dasyurus hallucatus) is an endangered carnivorous marsupial in the family Dasyuridae that occurs in the northern third of Australia. It has declined throughout its range, especially in open, lowland habitats. This led to the hypothesis that rocky outcrops where it persists provide a safe haven from introduced predators and provide greater microhabitat heterogeneity associated with higher prey availability. Proposed causes of decline include introduced cane toads (Rhinella marina, which are toxic), predation by feral cats (Felis catus), foxes (Vulpes vulpes), and the dingo (Canis lupus), and habitat alteration by changes in fire regimes. The National Recovery Plan highlighted knowledge gaps relevant to the conservation of the northern quoll. These include assembling data on ecology and population status, determining factors in survival, especially introduced predators, selecting areas that can be used as refuges, and identifying and securing key populations. The Pilbara region in Western Australia is a key population currently free of invasive cane toads (a major threat). The Pilbara is a semi-arid to arid area subject to cyclones between December and March. I selected two sites: Millstream Chichester National Park (2 381 km2) and Indee Station (1 623 km2). These are dominated by spinifex (hummock) grasslands, with rugged rock outcrops, shrublands, riparian areas, and some soft (tussock) grasslands. They are subject to frequent seasonal fires, creating a mosaic of recently burnt and longer unburnt areas. -
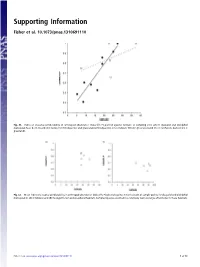
Supporting Information
Supporting Information Fisher et al. 10.1073/pnas.1310691110 Fig. S1. Index of seasonal predictability in arthropod abundance (Colwell’s P) plotted against latitude of sampling sites where dasyurid and didelphid marsupials have been recorded in rainforest (filled points) and grassland (unfilled points). Lines indicate fitted regressions (solid line = rainforest, dashed line = grassland). Fig. S2. Mean index of seasonal predictability in arthropod abundance (Colwell’s P) plotted against mean latitude of sample points for dasyurid and didelphid marsupials in (A) shrubland and (B) Eucalypt forest and woodland habitats. Sampled species occurred in a relatively narrow range of latitudes in these habitats. Fisher et al. www.pnas.org/cgi/content/short/1310691110 1of13 Fig. S3. Phylogeny of insectivorous marsupials with known life history data, based on ref. 1 with updates from ref. 2. 1. Cardillo M, Bininda-Emonds ORP, Boakes E, Purvis A (2004) A species-level phylogenetic supertree of marsupials. J Zool 264(1):11–31. 2. Fritz SA, Bininda-Emonds ORP, Purvis A (2009) Geographical variation in predictors of mammalian extinction risk: Big is bad, but only in the tropics. Ecol Lett 12(6):538–549. Fisher et al. www.pnas.org/cgi/content/short/1310691110 2of13 Fisher et al. Table S1. Reproductive traits and diets of insectivorous marsupials Latitude (south) www.pnas.org/cgi/content/short/1310691110 Proportion Proportion Female for Habitat of of age species class for females males Male Breeding Copulation Litters Scrotal at first with species Genus species -

BIO 597Z – Mammalogy Mammal Characteristics
BIO 597Z – Mammalogy • 4,629 species • Phylum Chordata "back-boned animals" Four Distinct Features 1) notocord = rod extending the length of the body; structural support 2) dorsal, hollow nerve cord located above notocord 3) pharynx, with gill slits 4) tail Mammal Characteristics • Subphylum Vertebrata Vertebrates = true back-bone - Vertebral column or backbone, generally replaces notocord - Brain enclosed in cranial cavity (e.g., skull) - Endoskeleton 1 Mammal Characteristics • Class Mammalia – Subclass Prototheria (monotremes) – Subclass Theria • Infraclass Metatheria (marsupials) • Infraclass Eutheria (placentals) Mammal Characteristics • Class Mammalia Distinguishing Features 1) Hair 2) Mammary glands (= milk for young) 3) Endothermic 4) Most bear live young* 5) Behavioral advancements over other classes, due to increased brain size/intelligence 2 Mammal Characteristics *Exception to the Rule: 1) monotremes: e.g., duck- billed platypus = egg laying mammal 2) marsupials: e.g., opossums, koala = bear live young which are immature; young develop in specialized pouch Mammals in Michigan 9 Orders of Mammals in MI – Didelphimorphia – Insectivora – Chiroptera –Primates – Carnivora – Perissodactyla – Artiodactyla – Lagomorpha – Rodentia 3 MAMMALIAN DIVERSITY: AN ORDERLY OVERVIEW A. Monotremes (Order Montremata) e.g, echidnas & duck-billed platypus MAMMALIAN DIVERSITY: AN ORDERLY OVERVIEW B. Marsupials 1. South American Marsupials a. Order Didelphimorphia Wooly opossum Virginia opossum Mouse opossum 4 MAMMALIAN DIVERSITY: AN ORDERLY OVERVIEW B. Marsupials 2. Australian Marsupials a. Order Dasyuromorphia Tasmanian devil MAMMALIAN DIVERSITY: AN ORDERLY OVERVIEW B. Marsupials 2. Australian Marsupials b. Order Peramelemorphia Rabbit-eared bandicoot 5 MAMMALIAN DIVERSITY: AN ORDERLY OVERVIEW B. Marsupials 2. Australian Marsupials c. Order Diprotodontia… C. Placental Mammals –Order Xenarthra (= Edentata) (anteaters, sloths, armadillos) Giant anteater Nine-banded armadillo 6 C. -

The Role of Feral Predators in Disrupting Small Vertebrate Communities in Arid South Australia
The role of feral predators in disrupting small vertebrate communities in arid South Australia Project Summary Project 1.1.4 Brush-tailed Mulgara in the Simpson Desert. Photo: Aaron Greenville Research in Brief Why is the research Strzelecki and Simpson Deserts. Lack of successful dispersal across This project is investigating why needed? non-refuge habitat types appears to native species persist in some Most medium-sized and many small be a limiting factor and will be the refuge areas of South Australia mammal species have fared poorly focus of this research. but not others, and the role of in the wake of early livestock grazing habitat condition and especially practices, successive rabbit plagues, How will the research feral predators in restricting and introduced predators. help? their populations. The kowari and fawn hopping mouse are The threatened kowari and fawn This project will test a series of threatened and other species hopping mouse suffered large range hypotheses about why native such as the plains mouse reductions following European species persist in some refuge areas and crest-tailed mulgara are occupation of Australia and now but not others, the role of habitat restricted in range. appear to be restricted to specific condition, and particularly predators, refuge habitat on vast stony plains in in restricting populations and All species of northern South north-east SA and south-west QLD. their dispersal. Australia are also at risk of fox The lack of shelter for larger predators, and cat predation, less so where such as the red fox and feral cat, and The project will contribute to a predator activity is suppressed further suppression by the dingo, much greater understanding of by dingoes, particularly on vast may work together in providing how predators (feral cat, fox and stony plains where cover is refuges for these species in this area.