Research Article
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Universidad De Concepción Facultad De Ciencias Naturales Y
Universidad de Concepción Facultad de Ciencias Naturales y Oceanográficas Programa de Magister en Ciencias mención Botánica DIVERSIDAD DE MACROHONGOS EN ÁREAS DESÉRTICAS DEL NORTE GRANDE DE CHILE Tesis presentada a la Facultad de Ciencias Naturales y Oceanográficas de la Universidad de Concepción para optar al grado de Magister en Ciencias mención Botánica POR: SANDRA CAROLINA TRONCOSO ALARCÓN Profesor Guía: Dr. Götz Palfner Profesora Co-Guía: Dra. Angélica Casanova Mayo 2020 Concepción, Chile AGRADECIMIENTOS Son muchas las personas que han contribuido durante el proceso y conclusión de este trabajo. En primer lugar quiero agradecer al Dr. Götz Palfner, guía de esta tesis y mi profesor desde el año 2014 y a la Dra. Angélica Casanova, quienes con su experiencia se han esforzado en enseñarme y ayudarme a llegar a esta instancia. A mis compañeros de laboratorio, Josefa Binimelis, Catalina Marín y Cristobal Araneda, por la ayuda mutua y compañía que nos pudimos brindar en el laboratorio o durante los viajes que se realizaron para contribuir a esta tesis. A CONAF Atacama y al Proyecto RT2716 “Ecofisiología de Líquenes Antárticos y del desierto de Atacama”, cuyas gestiones o financiamientos permitieron conocer un poco más de la diversidad de hongos y de los bellos paisajes de nuestro desierto chileno. Agradezco al Dr. Pablo Guerrero y a todas las personas que enviaron muestras fúngicas desertícolas al Laboratorio de Micología para identificarlas y que fueron incluidas en esta investigación. También agradezco a mi familia, mis padres, hermanos, abuelos y tíos, que siempre me han apoyado y animado a llevar a cabo mis metas de manera incondicional. -

A New Section and Two New Species of Podaxis (Gasteromycetes) from South Africa
S.Afr.l. Bot., 1989,55(2): 159-164 159 A new section and two new species of Podaxis (Gasteromycetes) from South Africa J.J.R. de Villiers, A. Eicker* and G.C.A. van der Westhuizen Department of Botany, University of Pretoria, Pretoria, 0001 Republic of South Africa Accepted 7 September /988 Podaxis africana De Villiers, Eicker & van der Westhuizen and P. rugospora De Villiers, Eicker & van der Westhuizen, two new species from Transvaal are described and illustrated. A new section, Umbricorticalis De Villiers, Eicker & van der Westhuizen is proposed in the genus Podaxis to accommodate P. africana. Morphologically P. africana resembles P. microporus McKnight but it is distinguished by its large globose, subglobose to broadly ovoid spores, the absence of a 'pin prick' pore structure, the black gleba, and the deep orange to strong brown inner cortex of the stipe. P. rugospora is allied to P. pistillaris (L. ex Pers.) Fr. emend. Morse from which it differs by reason of the hyaline, narrow, flattened, occasionally septate capillitium threads and the grayish-olive, light olive or light to moderate yellowish-brown gleba. The most remarkable character of these new species is the rugose spores. Podaxis africana De Villiers, Eicker & van der Westhuizen en P. rugospora De Villiers, Eicker & van der Westhuizen, twee nuwe spesies van Transvaal word beskryf en ge·illustreer. 'n Nuwe seksie, Umbricorticalis De Villiers, Eicker & van der Westhuizen in die genus Podaxis word voorgestel om P. africana te akkommodeer. Morfologies toon P. africana 'n ooreenkoms met P. microporus McKnight maar word deur die groot bolvormige, subbolvormige tot bykans eiervormige spore, die afwesigheid van 'n 'naaldprik'-porie struktuur, die swart gleba en die diep oranje tot sterk bruin binneste korteks van die stipe. -

Agaricales, Basidiomycota) Occurring in Punjab, India
Current Research in Environmental & Applied Mycology 5 (3): 213–247(2015) ISSN 2229-2225 www.creamjournal.org Article CREAM Copyright © 2015 Online Edition Doi 10.5943/cream/5/3/6 Ecology, Distribution Perspective, Economic Utility and Conservation of Coprophilous Agarics (Agaricales, Basidiomycota) Occurring in Punjab, India Amandeep K1*, Atri NS2 and Munruchi K2 1Desh Bhagat College of Education, Bardwal–Dhuri–148024, Punjab, India. 2Department of Botany, Punjabi University, Patiala–147002, Punjab, India. Amandeep K, Atri NS, Munruchi K 2015 – Ecology, Distribution Perspective, Economic Utility and Conservation of Coprophilous Agarics (Agaricales, Basidiomycota) Occurring in Punjab, India. Current Research in Environmental & Applied Mycology 5(3), 213–247, Doi 10.5943/cream/5/3/6 Abstract This paper includes the results of eco-taxonomic studies of coprophilous mushrooms in Punjab, India. The information is based on the survey to dung localities of the state during the various years from 2007-2011. A total number of 172 collections have been observed, growing as saprobes on dung of various domesticated and wild herbivorous animals in pastures, open areas, zoological parks, and on dung heaps along roadsides or along village ponds, etc. High coprophilous mushrooms’ diversity has been established and a number of rare and sensitive species recorded with the present study. The observed collections belong to 95 species spread over 20 genera and 07 families of the order Agaricales. The present paper discusses the distribution of these mushrooms in Punjab among different seasons, regions, habitats, and growing habits along with their economic utility, habitat management and conservation. This is the first attempt in which various dung localities of the state has been explored systematically to ascertain the diversity, seasonal availability, distribution and ecology of coprophilous mushrooms. -

Coprinus Comatus, a Newly Domesticated Wild Nutriceutical
Journal of Agricultural Technology 2009 Vol.5(2): 299-316 Journal of AgriculturalAvailable online Technology http://www.ijat-rmutto.com 2009, Vol.5(2): 299-316 ISSN 1686 -9141 Coprinus comatus , a newly domesticated wild nutriceutical mushroom in the Philippines Renato G. Reyes 1*, Lani Lou Mar A. Lopez 1, Kei Kumakura 2, Sofronio P. Kalaw 1, Tadahiro Kikukawa 3 and Fumio Eguchi 3 1Center for Tropical Mushroom Research and Development, College of Arts and Sciences, Central Luzon State University, Science City of Munoz, Nueva Ecija, Philippines 2Mush-Tech Co., Ltd., Japan 3Takasaki University of Health and Welfare, Gunma, Japan Reyes, R.G., Lopez, L.L.M.A., Kumakura, K., Kalaw, S.P., Kikukawa, T. and Eguchi, F. (2009). Coprinus comatus , a newly domesticated wild nutriceutical mushroom in the Philippines . Journal of Agricultural Technology 5(2): 299-316. The mycelial growth performance on indigenous culture media and the optimum physical conditions (pH, aeration and illumination) of C. comatus as a prelude to its domestication were investigated in this study. In our desire to develop technology for its aseptic cultivation for our immediate plan of using this mushroom for nutriceutical purposes, we have tried growing this mushroom under sterile condition. As such, the amino acid profile and toxicity of C. comatus were also elucidated. Results of our investigation revealed that C. comatus grown in sealed plates of coconut water gelatin (pH 6.5) produced very dense mycelial growth, 6 days after incubation in the dark. Early initiation of fruiting bodies was observed in bottles containing previously sterilized sawdust (8 parts): rice grit (2 parts) formulation. -
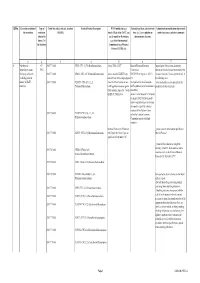
Qrno. 1 2 3 4 5 6 7 1 CP 2903 77 100 0 Cfcl3
QRNo. General description of Type of Tariff line code(s) affected, based on Detailed Product Description WTO Justification (e.g. National legal basis and entry into Administration, modification of previously the restriction restriction HS(2012) Article XX(g) of the GATT, etc.) force (i.e. Law, regulation or notified measures, and other comments (Symbol in and Grounds for Restriction, administrative decision) Annex 2 of e.g., Other International the Decision) Commitments (e.g. Montreal Protocol, CITES, etc) 12 3 4 5 6 7 1 Prohibition to CP 2903 77 100 0 CFCl3 (CFC-11) Trichlorofluoromethane Article XX(h) GATT Board of Eurasian Economic Import/export of these ozone destroying import/export ozone CP-X Commission substances from/to the customs territory of the destroying substances 2903 77 200 0 CF2Cl2 (CFC-12) Dichlorodifluoromethane Article 46 of the EAEU Treaty DECISION on August 16, 2012 N Eurasian Economic Union is permitted only in (excluding goods in dated 29 may 2014 and paragraphs 134 the following cases: transit) (all EAEU 2903 77 300 0 C2F3Cl3 (CFC-113) 1,1,2- 4 and 37 of the Protocol on non- On legal acts in the field of non- _to be used solely as a raw material for the countries) Trichlorotrifluoroethane tariff regulation measures against tariff regulation (as last amended at 2 production of other chemicals; third countries Annex No. 7 to the June 2016) EAEU of 29 May 2014 Annex 1 to the Decision N 134 dated 16 August 2012 Unit list of goods subject to prohibitions or restrictions on import or export by countries- members of the -

Diversity, Nutritional Composition and Medicinal Potential of Indian Mushrooms: a Review
Vol. 13(4), pp. 523-545, 22 January, 2014 DOI: 10.5897/AJB2013.13446 ISSN 1684-5315 ©2014 Academic Journals African Journal of Biotechnology http://www.academicjournals.org/AJB Review Diversity, nutritional composition and medicinal potential of Indian mushrooms: A review Hrudayanath Thatoi* and Sameer Kumar Singdevsachan Department of Biotechnology, College of Engineering and Technology, Biju Patnaik University of Technology, Bhubaneswar-751003, Odisha, India. Accepted 2 January, 2014 Mushrooms are the higher fungi which have long been used for food and medicinal purposes. They have rich nutritional value with high protein content (up to 44.93%), vitamins, minerals, fibers, trace elements and low calories and lack cholesterol. There are 14,000 known species of mushrooms of which 2,000 are safe for human consumption and about 650 of these possess medicinal properties. Among the total known mushrooms, approximately 850 species are recorded from India. Many of them have been used in food and folk medicine for thousands of years. Mushrooms are also sources of bioactive substances including antibacterial, antifungal, antiviral, antioxidant, antiinflammatory, anticancer, antitumour, anti-HIV and antidiabetic activities. Nutriceuticals and medicinal mushrooms have been used in human health development in India as food, medicine, minerals among others. The present review aims to update the current status of mushrooms diversity in India with their nutritional and medicinal potential as well as ethnomedicinal uses for different future prospects in pharmaceutical application. Key words: Mushroom diversity, nutritional value, therapeutic potential, bioactive compound. INTRODUCTION Mushroom is a general term used mainly for the fruiting unexamined mushrooms will be only 5%, implies that body of macrofungi (Ascomycota and Basidiomycota) there are 7,000 yet undiscovered species, which if and represents only a short reproductive stage in their life discovered will be provided with the possible benefit to cycle (Das, 2010). -

The Mycological Society of San Francisco • Jan. 2016, Vol. 67:05
The Mycological Society of San Francisco • Jan. 2016, vol. 67:05 Table of Contents JANUARY 19 General Meeting Speaker Mushroom of the Month by K. Litchfield 1 President Post by B. Wenck-Reilly 2 Robert Dale Rogers Schizophyllum by D. Arora & W. So 4 Culinary Corner by H. Lunan 5 Hospitality by E. Multhaup 5 Holiday Dinner 2015 Report by E. Multhaup 6 Bizarre World of Fungi: 1965 by B. Sommer 7 Academic Quadrant by J. Shay 8 Announcements / Events 9 2015 Fungus Fair by J. Shay 10 David Arora’s talk by D. Tighe 11 Cultivation Quarters by K. Litchfield 12 Fungus Fair Species list by D. Nolan 13 Calendar 15 Mushroom of the Month: Chanterelle by Ken Litchfield Twenty-One Myths of Medicinal Mushrooms: Information on the use of medicinal mushrooms for This month’s profiled mushroom is the delectable Chan- preventive and therapeutic modalities has increased terelle, one of the most distinctive and easily recognized mush- on the internet in the past decade. Some is based on rooms in all its many colors and meaty forms. These golden, yellow, science and most on marketing. This talk will look white, rosy, scarlet, purple, blue, and black cornucopias of succu- at 21 common misconceptions, helping separate fact lent brawn belong to the genera Cantharellus, Craterellus, Gomphus, from fiction. Turbinellus, and Polyozellus. Rather than popping up quickly from quiescent primordial buttons that only need enough rain to expand About the speaker: the preformed babies, Robert Dale Rogers has been an herbalist for over forty these mushrooms re- years. He has a Bachelor of Science from the Univer- quire an extended period sity of Alberta, where he is an assistant clinical profes- of slower growth and sor in Family Medicine. -

Fungi of North East Victoria Online
Agarics Agarics Agarics Agarics Fungi of North East Victoria An Identication and Conservation Guide North East Victoria encompasses an area of almost 20,000 km2, bounded by the Murray River to the north and east, the Great Dividing Range to the south and Fungi the Warby Ranges to the west. From box ironbark woodlands and heathy dry forests, open plains and wetlands, alpine herb elds, montane grasslands and of North East Victoria tall ash forests, to your local park or backyard, fungi are found throughout the region. Every fungus species contributes to the functioning, health and An Identification and Conservation Guide resilience of these ecosystems. Identifying Fungi This guide represents 96 species from hundreds, possibly thousands that grow in the diverse habitats of North East Victoria. It includes some of the more conspicuous and distinctive species that can be recognised in the eld, using features visible to the Agaricus xanthodermus* Armillaria luteobubalina* Coprinellus disseminatus Cortinarius austroalbidus Cortinarius sublargus Galerina patagonica gp* Hypholoma fasciculare Lepista nuda* Mycena albidofusca Mycena nargan* Protostropharia semiglobata Russula clelandii gp. yellow stainer Australian honey fungus fairy bonnet Australian white webcap funeral bell sulphur tuft blewit* white-crowned mycena Nargan’s bonnet dung roundhead naked eye or with a x10 magnier. LAMELLAE M LAMELLAE M ■ LAMELLAE S ■ LAMELLAE S, P ■ LAMELLAE S ■ LAMELLAE M ■ ■ LAMELLAE S ■ LAMELLAE S ■ LAMELLAE S ■ LAMELLAE S ■ LAMELLAE S ■ LAMELLAE S ■ When identifying a fungus, try and nd specimens of the same species at dierent growth stages, so you can observe the developmental changes that can occur. Also note the variation in colour and shape that can result from exposure to varying weather conditions. -

2011 NAMA Toxicology Committee Report North American Mushroom Poisonings
2011 NAMA Toxicology Committee Report North American Mushroom Poisonings By Michael W. Beug, PhD, Chair NAMA Toxicology Committee P.O. Box 116, Husum, WA 98623 email: [email protected] Abstract In 2011, for North America, the reports to NAMA included 117 people seriously sickened by mushrooms. Thirteen cases involved ingestion of either “Destroying Angel” or “Death Cap” mushrooms in the genus Amanita. There was one death and two people needed a liver transplant after ingestion of a “Destroying Angel”, presumably Amanita bisporigera. Three cases, including one death, involved amatoxins from a Galerina , presumably Galerina marginata . There was one case of kidney damage after consumption of Amanita smithiana and a second case of kidney damage involving two women who consumed an unknown mushroom . The year was noteworthy for the large number of reports of problems from consumption of morels with 22 cases (18.8% of the total). A number of problems were the result of people consuming morels raw – but everyone recovered within 24 hours. The eighteen Chlorophyllum molybdites cases (15% of the total) were sometimes quite severe and often required hospitalization. While Gyromitra cases numbered only nine (eight Gyromitra esculenta and one Gyromitra montana ), four required long hospitalizations as a result of liver damage. In the Northeast, newspaper reports mention long hospitalizations from some of the mushrooms known to cause gastro- intestinal distress, but we have no information on those cases. Twenty seven reports of dogs ill after eating mushrooms included fourteen deaths of the dogs. The dog deaths were mostly attributed to ingestion of mushrooms containing α-amanitin, including probable Amanita bisporigera, Amanita ocreata and Amanita phalloides , though in at least one case the possibility of a deadly Galerina cannot be ruled out. -

Postharvest Biochemical Characteristics and Ultrastructure of Coprinus Comatus
Postharvest biochemical characteristics and ultrastructure of Coprinus comatus Yi Peng1,2, Tongling Li2, Huaming Jiang3, Yunfu Gu1, Qiang Chen1, Cairong Yang2,4, Wei liang Qi2, Song-qing Liu2,4 and Xiaoping Zhang1 1 College of Resources, Sichuan Agricultural Uniersity, Chengdu, Sichuan, China 2 College of Chemistry and Life Sciences, Chengdu Normal University, Chengdu, Sichuan, China 3 Sichuan Vocational and Technical College, Suining, Sichuan, China 4 Institute of Microbiology, Chengdu Normal University, Chengdu, Sichuan, China ABSTRACT Background. Coprinus comatus is a novel cultivated edible fungus, hailed as a new preeminent breed of mushroom. However, C. comatus is difficult to keep fresh at room temperature after harvest due to high respiration, browning, self-dissolve and lack of physical protection. Methods. In order to extend the shelf life of C. comatus and reduce its loss in storage, changes in quality, biochemical content, cell wall metabolism and ultrastructure of C. comatus (C.c77) under 4 ◦C and 90% RH storage regimes were investigated in this study. Results. The results showed that: (1) After 10 days of storage, mushrooms appeared acutely browning, cap opening and flowing black juice, rendering the mushrooms commercially unacceptable. (2) The activity of SOD, CAT, POD gradually increased, peaked at the day 10, up to 31.62 U g−1 FW, 16.51 U g−1 FW, 0.33 U g−1 FW, respectively. High SOD, CAT, POD activity could be beneficial in protecting cells from ROS-induced injuries, alleviating lipid peroxidation and stabilizing membrane integrity. (3) The activities of chitinase, β-1,3-glucanase were significantly increased. Higher degrees of cell wall degradation observed during storage might be due to those enzymes' high activities. -

Fungal Diversity in the Mediterranean Area
Fungal Diversity in the Mediterranean Area • Giuseppe Venturella Fungal Diversity in the Mediterranean Area Edited by Giuseppe Venturella Printed Edition of the Special Issue Published in Diversity www.mdpi.com/journal/diversity Fungal Diversity in the Mediterranean Area Fungal Diversity in the Mediterranean Area Editor Giuseppe Venturella MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade • Manchester • Tokyo • Cluj • Tianjin Editor Giuseppe Venturella University of Palermo Italy Editorial Office MDPI St. Alban-Anlage 66 4052 Basel, Switzerland This is a reprint of articles from the Special Issue published online in the open access journal Diversity (ISSN 1424-2818) (available at: https://www.mdpi.com/journal/diversity/special issues/ fungal diversity). For citation purposes, cite each article independently as indicated on the article page online and as indicated below: LastName, A.A.; LastName, B.B.; LastName, C.C. Article Title. Journal Name Year, Article Number, Page Range. ISBN 978-3-03936-978-2 (Hbk) ISBN 978-3-03936-979-9 (PDF) c 2020 by the authors. Articles in this book are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book as a whole is distributed by MDPI under the terms and conditions of the Creative Commons license CC BY-NC-ND. Contents About the Editor .............................................. vii Giuseppe Venturella Fungal Diversity in the Mediterranean Area Reprinted from: Diversity 2020, 12, 253, doi:10.3390/d12060253 .................... 1 Elias Polemis, Vassiliki Fryssouli, Vassileios Daskalopoulos and Georgios I. -

Mantar Dergisi
11 6845 - Volume: 20 Issue:1 JOURNAL - E ISSN:2147 - April 20 e TURKEY - KONYA - FUNGUS Research Center JOURNAL OF OF JOURNAL Selçuk Selçuk University Mushroom Application and Selçuk Üniversitesi Mantarcılık Uygulama ve Araştırma Merkezi KONYA-TÜRKİYE MANTAR DERGİSİ E-DERGİ/ e-ISSN:2147-6845 Nisan 2020 Cilt:11 Sayı:1 e-ISSN 2147-6845 Nisan 2020 / Cilt:11/ Sayı:1 April 2020 / Volume:11 / Issue:1 SELÇUK ÜNİVERSİTESİ MANTARCILIK UYGULAMA VE ARAŞTIRMA MERKEZİ MÜDÜRLÜĞÜ ADINA SAHİBİ PROF.DR. GIYASETTİN KAŞIK YAZI İŞLERİ MÜDÜRÜ DR. ÖĞR. ÜYESİ SİNAN ALKAN Haberleşme/Correspondence S.Ü. Mantarcılık Uygulama ve Araştırma Merkezi Müdürlüğü Alaaddin Keykubat Yerleşkesi, Fen Fakültesi B Blok, Zemin Kat-42079/Selçuklu-KONYA Tel:(+90)0 332 2233998/ Fax: (+90)0 332 241 24 99 Web: http://mantarcilik.selcuk.edu.tr http://dergipark.gov.tr/mantar E-Posta:[email protected] Yayın Tarihi/Publication Date 27/04/2020 i e-ISSN 2147-6845 Nisan 2020 / Cilt:11/ Sayı:1 / / April 2020 Volume:11 Issue:1 EDİTÖRLER KURULU / EDITORIAL BOARD Prof.Dr. Abdullah KAYA (Karamanoğlu Mehmetbey Üniv.-Karaman) Prof.Dr. Abdulnasır YILDIZ (Dicle Üniv.-Diyarbakır) Prof.Dr. Abdurrahman Usame TAMER (Celal Bayar Üniv.-Manisa) Prof.Dr. Ahmet ASAN (Trakya Üniv.-Edirne) Prof.Dr. Ali ARSLAN (Yüzüncü Yıl Üniv.-Van) Prof.Dr. Aysun PEKŞEN (19 Mayıs Üniv.-Samsun) Prof.Dr. A.Dilek AZAZ (Balıkesir Üniv.-Balıkesir) Prof.Dr. Ayşen ÖZDEMİR TÜRK (Anadolu Üniv.- Eskişehir) Prof.Dr. Beyza ENER (Uludağ Üniv.Bursa) Prof.Dr. Cvetomir M. DENCHEV (Bulgarian Academy of Sciences, Bulgaristan) Prof.Dr. Celaleddin ÖZTÜRK (Selçuk Üniv.-Konya) Prof.Dr. Ertuğrul SESLİ (Trabzon Üniv.-Trabzon) Prof.Dr.