Shaffer2009chap55.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Egyptian Tortoise (Testudo Kleinmanni)
EAZA Reptile Taxon Advisory Group Best Practice Guidelines for the Egyptian tortoise (Testudo kleinmanni) First edition, May 2019 Editors: Mark de Boer, Lotte Jansen & Job Stumpel EAZA Reptile TAG chair: Ivan Rehak, Prague Zoo. EAZA Best Practice Guidelines Egyptian tortoise (Testudo kleinmanni) EAZA Best Practice Guidelines disclaimer Copyright (May 2019) by EAZA Executive Office, Amsterdam. All rights reserved. No part of this publication may be reproduced in hard copy, machine-readable or other forms without advance written permission from the European Association of Zoos and Aquaria (EAZA). Members of the European Association of Zoos and Aquaria (EAZA) may copy this information for their own use as needed. The information contained in these EAZA Best Practice Guidelines has been obtained from numerous sources believed to be reliable. EAZA and the EAZA Reptile TAG make a diligent effort to provide a complete and accurate representation of the data in its reports, publications, and services. However, EAZA does not guarantee the accuracy, adequacy, or completeness of any information. EAZA disclaims all liability for errors or omissions that may exist and shall not be liable for any incidental, consequential, or other damages (whether resulting from negligence or otherwise) including, without limitation, exemplary damages or lost profits arising out of or in connection with the use of this publication. Because the technical information provided in the EAZA Best Practice Guidelines can easily be misread or misinterpreted unless properly analysed, EAZA strongly recommends that users of this information consult with the editors in all matters related to data analysis and interpretation. EAZA Preamble Right from the very beginning it has been the concern of EAZA and the EEPs to encourage and promote the highest possible standards for husbandry of zoo and aquarium animals. -
N.C. Turtles Checklist
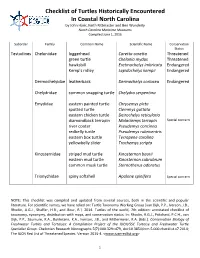
Checklist of Turtles Historically Encountered In Coastal North Carolina by John Hairr, Keith Rittmaster and Ben Wunderly North Carolina Maritime Museums Compiled June 1, 2016 Suborder Family Common Name Scientific Name Conservation Status Testudines Cheloniidae loggerhead Caretta caretta Threatened green turtle Chelonia mydas Threatened hawksbill Eretmochelys imbricata Endangered Kemp’s ridley Lepidochelys kempii Endangered Dermochelyidae leatherback Dermochelys coriacea Endangered Chelydridae common snapping turtle Chelydra serpentina Emydidae eastern painted turtle Chrysemys picta spotted turtle Clemmys guttata eastern chicken turtle Deirochelys reticularia diamondback terrapin Malaclemys terrapin Special concern river cooter Pseudemys concinna redbelly turtle Pseudemys rubriventris eastern box turtle Terrapene carolina yellowbelly slider Trachemys scripta Kinosternidae striped mud turtle Kinosternon baurii eastern mud turtle Kinosternon subrubrum common musk turtle Sternotherus odoratus Trionychidae spiny softshell Apalone spinifera Special concern NOTE: This checklist was compiled and updated from several sources, both in the scientific and popular literature. For scientific names, we have relied on: Turtle Taxonomy Working Group [van Dijk, P.P., Iverson, J.B., Rhodin, A.G.J., Shaffer, H.B., and Bour, R.]. 2014. Turtles of the world, 7th edition: annotated checklist of taxonomy, synonymy, distribution with maps, and conservation status. In: Rhodin, A.G.J., Pritchard, P.C.H., van Dijk, P.P., Saumure, R.A., Buhlmann, K.A., Iverson, J.B., and Mittermeier, R.A. (Eds.). Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Chelonian Research Monographs 5(7):000.329–479, doi:10.3854/crm.5.000.checklist.v7.2014; The IUCN Red List of Threatened Species. -

A Phylogenomic Analysis of Turtles ⇑ Nicholas G
Molecular Phylogenetics and Evolution 83 (2015) 250–257 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev A phylogenomic analysis of turtles ⇑ Nicholas G. Crawford a,b,1, James F. Parham c, ,1, Anna B. Sellas a, Brant C. Faircloth d, Travis C. Glenn e, Theodore J. Papenfuss f, James B. Henderson a, Madison H. Hansen a,g, W. Brian Simison a a Center for Comparative Genomics, California Academy of Sciences, 55 Music Concourse Drive, San Francisco, CA 94118, USA b Department of Genetics, University of Pennsylvania, Philadelphia, PA 19104, USA c John D. Cooper Archaeological and Paleontological Center, Department of Geological Sciences, California State University, Fullerton, CA 92834, USA d Department of Biological Sciences, Louisiana State University, Baton Rouge, LA 70803, USA e Department of Environmental Health Science, University of Georgia, Athens, GA 30602, USA f Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA g Mathematical and Computational Biology Department, Harvey Mudd College, 301 Platt Boulevard, Claremont, CA 9171, USA article info abstract Article history: Molecular analyses of turtle relationships have overturned prevailing morphological hypotheses and Received 11 July 2014 prompted the development of a new taxonomy. Here we provide the first genome-scale analysis of turtle Revised 16 October 2014 phylogeny. We sequenced 2381 ultraconserved element (UCE) loci representing a total of 1,718,154 bp of Accepted 28 October 2014 aligned sequence. Our sampling includes 32 turtle taxa representing all 14 recognized turtle families and Available online 4 November 2014 an additional six outgroups. Maximum likelihood, Bayesian, and species tree methods produce a single resolved phylogeny. -

The Conservation Biology of Tortoises
The Conservation Biology of Tortoises Edited by Ian R. Swingland and Michael W. Klemens IUCN/SSC Tortoise and Freshwater Turtle Specialist Group and The Durrell Institute of Conservation and Ecology Occasional Papers of the IUCN Species Survival Commission (SSC) No. 5 IUCN—The World Conservation Union IUCN Species Survival Commission Role of the SSC 3. To cooperate with the World Conservation Monitoring Centre (WCMC) The Species Survival Commission (SSC) is IUCN's primary source of the in developing and evaluating a data base on the status of and trade in wild scientific and technical information required for the maintenance of biological flora and fauna, and to provide policy guidance to WCMC. diversity through the conservation of endangered and vulnerable species of 4. To provide advice, information, and expertise to the Secretariat of the fauna and flora, whilst recommending and promoting measures for their con- Convention on International Trade in Endangered Species of Wild Fauna servation, and for the management of other species of conservation concern. and Flora (CITES) and other international agreements affecting conser- Its objective is to mobilize action to prevent the extinction of species, sub- vation of species or biological diversity. species, and discrete populations of fauna and flora, thereby not only maintain- 5. To carry out specific tasks on behalf of the Union, including: ing biological diversity but improving the status of endangered and vulnerable species. • coordination of a programme of activities for the conservation of biological diversity within the framework of the IUCN Conserva- tion Programme. Objectives of the SSC • promotion of the maintenance of biological diversity by monitor- 1. -

The Common Snapping Turtle, Chelydra Serpentina
The Common Snapping Turtle, Chelydra serpentina Rylen Nakama FISH 423: Olden 12/5/14 Figure 1. The Common Snapping Turtle, one of the most widespread reptiles in North America. Photo taken in Quebec, Canada. Image from https://www.flickr.com/photos/yorthopia/7626614760/. Classification Order: Testudines Family: Chelydridae Genus: Chelydra Species: serpentina (Linnaeus, 1758) Previous research on Chelydra serpentina (Phillips et al., 1996) acknowledged four subspecies, C. s. serpentina (Northern U.S. and Figure 2. Side profile of Chelydra serpentina. Note Canada), C. s. osceola (Southeastern U.S.), C. s. the serrated posterior end of the carapace and the rossignonii (Central America), and C. s. tail’s raised central ridge. Photo from http://pelotes.jea.com/AnimalFact/Reptile/snapturt.ht acutirostris (South America). Recent IUCN m. reclassification of chelonians based on genetic analyses (Rhodin et al., 2010) elevated C. s. rossignonii and C. s. acutirostris to species level and established C. s. osceola as a synonym for C. s. serpentina, thus eliminating subspecies within C. serpentina. Antiquated distinctions between the two formerly recognized North American subspecies were based on negligible morphometric variations between the two populations. Interbreeding in the overlapping range of the two populations was well documented, further discrediting the validity of the subspecies distinction (Feuer, 1971; Aresco and Gunzburger, 2007). Therefore, any emphasis of subspecies differentiation in the ensuing literature should be disregarded. Figure 3. Front-view of a captured Chelydra Continued usage of invalid subspecies names is serpentina. Different skin textures and the distinctive pink mouth are visible from this angle. Photo from still prevalent in the exotic pet trade for C. -

Dermatemys Mawii (The Hicatee, Tortuga Blanca, Or Central American River Turtle): a Working Bibliography
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/325206006 Dermatemys mawii (The Hicatee, Tortuga Blanca, or Central American River Turtle): A Working Bibliography Article · May 2018 CITATIONS READS 0 521 6 authors, including: Venetia Briggs Sergio C. Gonzalez University of Florida University of Florida 16 PUBLICATIONS 133 CITATIONS 9 PUBLICATIONS 39 CITATIONS SEE PROFILE SEE PROFILE Thomas Rainwater Clemson University 143 PUBLICATIONS 1,827 CITATIONS SEE PROFILE Some of the authors of this publication are also working on these related projects: Crocodiles in the Everglades View project Dissertation: The role of habitat expansion on amphibian community structure View project All content following this page was uploaded by Sergio C. Gonzalez on 17 May 2018. The user has requested enhancement of the downloaded file. 2018 Endangered and ThreatenedCaribbean Species Naturalist of the Caribbean Region Special Issue No. 2 V. Briggs-Gonzalez, S.C. Gonzalez, D. Smith, K. Allen, T.R. Rainwater, and F.J. Mazzotti 2018 CARIBBEAN NATURALIST Special Issue No. 2:1–22 Dermatemys mawii (The Hicatee, Tortuga Blanca, or Central American River Turtle): A Working Bibliography Venetia Briggs-Gonzalez1,*, Sergio C. Gonzalez1, Dustin Smith2, Kyle Allen1, Thomas R. Rainwater3, and Frank J. Mazzotti1 Abstract - Dermatemys mawii (Central American River Turtle), locally known in Belize as the “Hicatee” and in Guatemala and Mexico as Tortuga Blanca, is a large, highly aquatic freshwater turtle that has been extirpated from much of its historical range of southern Mex- ico, northern Guatemala, and lowland Belize. Throughout its restricted range, Dermatemys has been intensely harvested for its meat and eggs and sold in local markets. -

Mitochondrial Genome Reveals Contrasting Pattern of Adaptive
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.18.431795; this version posted February 18, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. MITOCHONDRIAL GENOME REVEALS CONTRASTING PATTERN OF ADAPTIVE SELECTION IN TURTLES AND TORTOISES Subhashree Sahoo1,2, Ajit Kumar1, Jagdish Rai2, Sandeep Kumar Gupta1 1. Department of Animal Ecology and Conservation Biology, Wildlife Institute of India, Dehradun 2. Institute of Forensic Science and Criminology, Panjab University, India *Author for correspondence Dr. S.K. Gupta Scientist-E Wildlife Institute of India Dehra Dun 248 001, India E-mail: [email protected] Telephone: +91-135-2646343 Fax No.: +91-135-2640117 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.02.18.431795; this version posted February 18, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Testudinoidea represents an evolutionarily unique taxon comprising both turtles and tortoises. The contrasting habitats that turtles and tortoises inhabit are associated with unique physio-ecological challenges hence enable distinct adaptive evolutionary strategies. To comparatively understand the pattern and strength of Darwinian selection and physicochemical evolution in turtle and tortoise mitogenomes, we employed adaptive divergence and selection analyses. We evaluated changes in structural and biochemical properties, and codon models on the mitochondrial protein-coding genes (PCGs) among three turtles and a tortoise lineage. -

Ecol 483/583 – Herpetology Lab 11: Reptile Diversity 3: Testudines and Crocodylia Spring 2010
Ecol 483/583 – Herpetology Lab 11: Reptile Diversity 3: Testudines and Crocodylia Spring 2010 P.J. Bergmann & S. Foldi Lab objectives The objectives of today’s lab are to: 1. Familiarize yourselves with extant diversity of the Testudines and Crocodylia. 2. Learn to identify species of Testundines that live in Arizona. Today's lab is the final lab on "reptile" diversity, and will introduce you to the Testudines, or turtles and tortoises, Crocodylia. Although there are no crocodylians in Arizona and the Testudine diversity is lower than that of the lizards or snakes, there is still a fair amount of material to learn, so use your time wisely. Tips for learning the material At this point in the course, your skills for learning herp diversity should be well honed, so continue with the strategies you have already learned during the semester. Learn how to differentiate the three major clades of Crocodylians, and the more numerous Testudine clades. There is another keying exercise in this lab, focusing on the Testudines. 1 Ecol 483/583 – Lab 11: Testudines & Crocs 2010 Exercise 1: Testudines (Modified from Bonine & Foldi 2008; Bonine, Dee & Hall 2006; Edwards 2002; Prival 2000) General information Turtles are probably the most instantly recognizable groups of all reptiles because of their shell and the ability to withdraw their heads and limbs into this protective structure. Turtles are a monophyletic group comprising the order Testudines also called Chelonia . Testudines is the term used to denote all members of the order (extant and extinct) whereas Chelonia is often used to denote extant turtles. -

The Natural History & Distribution of Riverine Turtles in West Virginia
Marshall University Marshall Digital Scholar Theses, Dissertations and Capstones 2010 The aN tural History & Distribution of Riverine Turtles in West Virginia Linh Diem Phu Follow this and additional works at: http://mds.marshall.edu/etd Part of the Aquaculture and Fisheries Commons, and the Terrestrial and Aquatic Ecology Commons Recommended Citation Phu, Linh Diem, "The aN tural History & Distribution of Riverine Turtles in West Virginia" (2010). Theses, Dissertations and Capstones. Paper 787. This Thesis is brought to you for free and open access by Marshall Digital Scholar. It has been accepted for inclusion in Theses, Dissertations and Capstones by an authorized administrator of Marshall Digital Scholar. For more information, please contact [email protected]. The Natural History & Distribution of Riverine Turtles in West Virginia Thesis submitted to the Graduate College of Marshall University In partial fulfillment of the requirements for the degree of Master of Science in Biological Sciences By Linh Diem Phu Dr. Thomas K. Pauley, Ph.D., Committee Chairperson Dr. Dan Evans, Ph.D. Dr. Suzanne Strait, Ph.D. Marshall University May 2010 Abstract Turtles are unique evolutionary marvels that evolved from amphibians and developed their protective shelled form more than 200 million years ago. In West Virginia, there are 10 native species of turtles, 9 of which are aquatic. Most of these aquatic turtles feed on carrion and dead plant matter, in the water and essentially "clean" our water systems. Turtles are long-lived animals with sensitive life stages that can serve as both long-term and short-term bioindicators of environmental health. With the increase in commercial trade, habitat fragmentation, degradation, destruction, there has been a marked decline in turtle species. -

Effect of Rainfall on Loggerhead Turtle Nest Temperatures, Sand Temperatures and Hatchling Sex
Vol. 28: 235–247, 2015 ENDANGERED SPECIES RESEARCH Published online October 7 doi: 10.3354/esr00684 Endang Species Res OPENPEN ACCESSCCESS Effect of rainfall on loggerhead turtle nest temperatures, sand temperatures and hatchling sex Alexandra Lolavar, Jeanette Wyneken* Department of Biological Sciences, Florida Atlantic University, 777 Glades Rd, Boca Raton, FL, USA ABSTRACT: Marine turtles deposit their eggs in underground nests where they develop unattended and without parental care. Incubation temperature varies with environmental conditions, including rainfall, sun/shade and sand type, and affects developmental rates, hatch and emergence success, and embryonic sex. We documented (1) rainfall and sand temperature relationships and (2) rainfall, nest temperatures and hatchling sex ratios at a loggerhead turtle (Caretta caretta) nesting beach in Boca Raton, Florida, USA, across the 2010 to 2013 nesting seasons. Rainfall data collected concur- rently with sand temperatures at different depths showed that light rainfall affected surface sand; effects of the heaviest rainfall events tended to lower sand temperatures but the temperature fluctu- ations were small once upper nest depths were reached. This is important in understanding the po- tential impacts of rainfall as a modifier of nest temperatures, as such changes can be quite small. Nest temperature profiles were synchronized with rainfall data from weather services to identify re- lationships with hatchling sex ratios. The sex of each turtle was verified laparoscopically to provide empirical measures of sex ratio for the nest and nesting beach. The majority of hatchlings in the samples were female, suggesting that across the 4 seasons most nest temperatures were not suffi- ciently cool to produce males. -

Conservation of South African Tortoises with Emphasis on Their Apicomplexan Haematozoans, As Well As Biological and Metal-Fingerprinting of Captive Individuals
CONSERVATION OF SOUTH AFRICAN TORTOISES WITH EMPHASIS ON THEIR APICOMPLEXAN HAEMATOZOANS, AS WELL AS BIOLOGICAL AND METAL-FINGERPRINTING OF CAPTIVE INDIVIDUALS By Courtney Antonia Cook THESIS submitted in fulfilment of the requirements for the degree PHILOSOPHIAE DOCTOR (Ph.D.) in ZOOLOGY in the FACULTY OF SCIENCE at the UNIVERSITY OF JOHANNESBURG Supervisor: Prof. N. J. Smit Co-supervisors: Prof. A. J. Davies and Prof. V. Wepener June 2012 “We need another and a wiser and perhaps a more mystical concept of animals. Remote from universal nature, and living by complicated artifice, man in civilization surveys the creature through the glass of his knowledge and sees thereby a feather magnified and the whole image in distortion. We patronize them for their incompleteness, for their tragic fate of having taken form so far below ourselves. And therein we err, and greatly err. For the animal shall not be measured by man. In a world older and more complete than ours they move finished and complete, gifted with extensions of the senses we have lost or never attained, living by voices we shall never hear. They are not brethren, they are not underlings; they are other nations caught with ourselves in the net of life and time, fellow prisoners of the splendour and travail of the earth.” Henry Beston (1928) ABSTRACT South Africa has the highest biodiversity of tortoises in the world with possibly an equivalent diversity of apicomplexan haematozoans, which to date have not been adequately researched. Prior to this study, five apicomplexans had been recorded infecting southern African tortoises, including two haemogregarines, Haemogregarina fitzsimonsi and Haemogregarina parvula, and three haemoproteids, Haemoproteus testudinalis, Haemoproteus balazuci and Haemoproteus sp. -

A Field Guide to South Dakota Turtles
A Field Guide to SOUTH DAKOTA TURTLES EC919 South Dakota State University | Cooperative Extension Service | USDA U.S. Geological Survey | South Dakota Cooperative Fish and Wildlife Research Unit South Dakota Department of Game, Fish & Parks This publication may be cited as: Bandas, Sarah J., and Kenneth F. Higgins. 2004. Field Guide to South Dakota Turtles. SDCES EC 919. Brookings: South Dakota State University. Copies may be obtained from: Dept. of Wildlife & Fisheries Sciences South Dakota State University Box 2140B, NPBL Brookings SD 57007-1696 South Dakota Dept of Game, Fish & Parks 523 E. Capitol, Foss Bldg Pierre SD 57501 SDSU Bulletin Room ACC Box 2231 Brookings, SD 57007 (605) 688–4187 A Field Guide to SOUTH DAKOTA TURTLES EC919 South Dakota State University | Cooperative Extension Service | USDA U.S. Geological Survey | South Dakota Cooperative Fish and Wildlife Research Unit South Dakota Department of Game, Fish & Parks Sarah J. Bandas Department of Wildlife and Fisheries Sciences South Dakota State University NPB Box 2140B Brookings, SD 57007 Kenneth F. Higgins U.S. Geological Survey South Dakota Cooperative Fish and Wildlife Research Unit South Dakota State University NPB Box 2140B Brookings, SD 57007 Contents 2 Introduction . .3 Status of South Dakota turtles . .3 Fossil record and evolution . .4 General turtle information . .4 Taxonomy of South Dakota turtles . .9 Capturing techniques . .10 Turtle handling . .10 Turtle habitats . .13 Western Painted Turtle (Chrysemys picta bellii) . .15 Snapping Turtle (Chelydra serpentina) . .17 Spiny Softshell Turtle (Apalone spinifera) . .19 Smooth Softshell Turtle (Apalone mutica) . .23 False Map Turtle (Graptemys pseudogeographica) . .25 Western Ornate Box Turtle (Terrapene ornata ornata) .