Enzymatic Properties of Human Kallikrein-Related Peptidase 12 (KLK12)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human Kallikrein Gene 11 (KLK11) Mrna Overexpression Is Associated with Poor Prognosis in Patients with Epithelial Ovarian Cancer
2766 Vol. 10, 2766–2770, April 15, 2004 Clinical Cancer Research Human Kallikrein Gene 11 (KLK11) mRNA Overexpression Is Associated with Poor Prognosis in Patients with Epithelial Ovarian Cancer 0.0225 ؍ Kazushi Shigemasa,1 Lijun Gu,1 significantly associated with overall survival (P ؍ Hirotoshi Tanimoto,2 Timothy J. O’Brien,3 and and P 0.0202, respectively) after multivariate analysis. Conclusions: KLK11 expression may play an important Koso Ohama1 role in ovarian cancer development and act as an independ- 1 Department of Obstetrics and Gynecology, Hiroshima University ent prognostic marker in ovarian cancer patients. Graduate School of Biomedical Sciences, Hiroshima, Japan; 2Department of Gynecology, National Hiroshima Hospital, Higashi Hiroshima, Japan; and 3Departments of Biochemistry and Molecular INTRODUCTION Biology and Obstetrics and Gynecology, University of Arkansas for Serine proteases comprise a family of protein-degrading Medical Sciences, Little Rock, Arkansas enzymes that serve a variety of biological functions, including induction of blood coagulation, activation of growth and angio- ABSTRACT genic factors, and degradation of extracellular matrix compo- nents (1–4). Purpose: The purpose of this study was to examine The human kallikrein gene family, a subfamily of serine expression levels of the human tissue kallikrein 11 gene proteases, is located at the chromosomal locus 19q13.3-q13.4. (KLK11) in epithelial ovarian tumors and to identify the Until recently, this family was thought to include only three relationship between KLK11 expression and patient sur- genes, the pancreatic renal kallikrein gene (KLK1), the human vival. glandular kallikrein gene (KLK2), and KLK3, which encodes Experimental Design: KLK11 mRNA expression was prostate-specific antigen (hK3). -

An Overview of the Kallikrein Gene Families in Humans and Other Species: Emerging Candidate Tumour Markers૾
Clinical Biochemistry 36 (2003) 443–452 An overview of the kallikrein gene families in humans and other species: Emerging candidate tumour markers૾ George M. Yousefa,b, Eleftherios P. Diamandisa,b,* aDepartment of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Ontario, Canada bDepartment of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Ontario, Canada Abstract Kallikreins are serine proteases with diverse physiologic functions. They are represented by multigene families in many animal species, especially in rat and mouse. Recently, the human kallikrein gene family has been fully characterized and includes 15 members, tandemly localized on chromosome 19q13.4. A new definition has now been proposed for kallikreins, which is not based on function but, rather, on close proximity and structural similarities. In this review, we summarize available information about kallikreins in many animal species with special emphasis on human kallikreins. We discuss the common structural features of kallikreins at the DNA, mRNA and protein levels and overview their evolutionary history. Kallikreins are expressed in a wide range of tissues including the salivary gland, endocrine or endocrine-related tissues such as testis, prostate, breast and endometrium and in the central nervous system. Most, if not all, genes are under steroid hormone regulation. Accumulating evidence indicates that kallikreins are involved in many pathologic conditions. Of special interest is the potential role of kallikreins in the central nervous system. In addition, many kallikreins seem to be candidate tumor markers for many malignancies, especially those of endocrine-related organs. © 2003 The Canadian Society of Clinical Chemists. All rights reserved. Keywords: Kallikrein; Tumor markers; Cancer biomarkers; Prostate cancer; Breast cancer; Ovarian cancer; Alzheimer’s disease; Serine proteases; Chromosome 19; Kallikrein evolution; Rodent kallikreins; Hormonally regulated genes 1. -

Kallikrein 13: a New Player in Coronaviral Infections
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.01.971499; this version posted March 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Kallikrein 13: a new player in coronaviral infections. 2 3 Aleksandra Milewska1,2, Katherine Falkowski2, Magdalena Kalinska3, Ewa Bielecka3, 4 Antonina Naskalska1, Pawel Mak4, Adam Lesner5, Marek Ochman6, Maciej Urlik6, Jan 5 Potempa2,7, Tomasz Kantyka3,8, Krzysztof Pyrc1,* 6 7 1 Virogenetics Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian 8 University, Gronostajowa 7a, 30-387 Krakow, Poland. 9 2 Microbiology Department, Faculty of Biochemistry, Biophysics and Biotechnology, 10 Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland. 11 3 Laboratory of Proteolysis and Post-translational Modification of Proteins, Malopolska 12 Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, 13 Poland. 14 4 Department of Analytical Biochemistry, Faculty of Biochemistry, Biophysics and 15 Biotechnology, Jagiellonian University, Gronostajowa 7 St., 30-387, Krakow, Poland. 16 5 University of Gdansk, Faculty of Chemistry, Wita Stwosza 63, 80-308 Gdansk, Poland. 17 6 Department of Cardiac, Vascular and Endovascular Surgery and Transplantology, Medical 18 University of Silesia in Katowice, Silesian Centre for Heart Diseases, Zabrze, Poland. 19 7 Centre for Oral Health and Systemic Diseases, University of Louisville School of Dentistry, 20 Louisville, KY 40202, USA. 21 8 Broegelmann Research Laboratory, Department of Clinical Science, University of Bergen, 22 5020 Bergen, Norway 23 24 25 26 27 28 29 30 31 * Correspondence should be addressed to Krzysztof Pyrc ([email protected]), Virogenetics 32 Laboratory of Virology, Malopolska Centre of Biotechnology, Jagiellonian University, 33 Gronostajowa 7, 30-387 Krakow, Poland; Phone number: +48 12 664 61 21; www: 34 http://virogenetics.info/. -
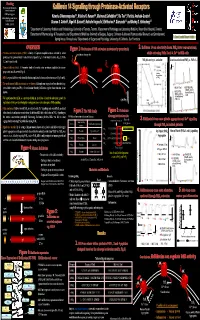
AACR2006-1686P
Funding Proteinases and Inflammation Network Group grant Kallikrein 14 Signalling through Proteinase-Activated Receptors CIHR operating grant 1,2 3,4 3,4 3,4 5 Alberta Heritage Foundation for Katerina Oikonomopoulou , Kristina K. Hansen , Mahmoud Saifeddine , Illa Tea , Patricia Andrade-Gordon , Medical Research Graeme S. Cottrell6, Nigel W. Bunnett6, Nathalie Vergnolle3, Eleftherios P. Diamandis1,2 and Morley D. Hollenberg3,4 NSERC / CRSNG 1Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto; 2Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto; 3Department of Pharmacology & Therapeutics, and 4Department of Medicine, University of Calgary, Calgary; 5Johnson & Johnson Pharmaceutical Research and Development, Spring House, Pennsylvania, 6Departments of Surgery and Physiology, University of California, San Francisco Kallikrein 14 can selectively disarm PAR (lower concentrations), OVERVIEW Figure 1: Mechanism of PAR activation (activation by proteolysis) 2. Kallikrein 14 can selectively disarm PAR1 (lower concentrations), 2+ Proteinase-activated receptors (PARs): a family of G-protein coupled receptors activated by serine proteinase cleavage site whilst activating PARs 2 and 4; Ca in HEK cells proteinases via a proteolytically revealed ‘tethered ligand’ (Fig. 1). Four family members (Fig. 2); PARs N PAR disarming 1, 2 and 4 signal to cells. N PAR1 dis-arming1 vs. activation Selective activation of PAR4 vs. PARs 1, 2 Human kallikreins (hKs): A 15-member family of secreted serine proteinases implicated in tumour 1 cm Thrombin 1 cm 2 min 2 min TFLLR-NH2 progression and cell survival (Fig. 4). (selective desensitization hK14: a tryptic kallikrein; wide tissue distribution, implicated in breast and ovarian cancer (Fig. 5 and 6). of PAR1) The mechanism of kallikrein action is not yet known: Although some targets have been identified (e.g. -

1 No. Affymetrix ID Gene Symbol Genedescription Gotermsbp Q Value 1. 209351 at KRT14 Keratin 14 Structural Constituent of Cyto
1 Affymetrix Gene Q No. GeneDescription GOTermsBP ID Symbol value structural constituent of cytoskeleton, intermediate 1. 209351_at KRT14 keratin 14 filament, epidermis development <0.01 biological process unknown, S100 calcium binding calcium ion binding, cellular 2. 204268_at S100A2 protein A2 component unknown <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 3. 33323_r_at SFN stratifin/14-3-3σ binding <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 4. 33322_i_at SFN stratifin/14-3-3σ binding <0.01 structural constituent of cytoskeleton, intermediate 5. 201820_at KRT5 keratin 5 filament, epidermis development <0.01 structural constituent of cytoskeleton, intermediate 6. 209125_at KRT6A keratin 6A filament, ectoderm development <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 7. 209260_at SFN stratifin/14-3-3σ binding <0.01 structural constituent of cytoskeleton, intermediate 8. 213680_at KRT6B keratin 6B filament, ectoderm development <0.01 receptor activity, cytosol, integral to plasma membrane, cell surface receptor linked signal transduction, sensory perception, tumor-associated calcium visual perception, cell 9. 202286_s_at TACSTD2 signal transducer 2 proliferation, membrane <0.01 structural constituent of cytoskeleton, cytoskeleton, intermediate filament, cell-cell adherens junction, epidermis 10. 200606_at DSP desmoplakin development <0.01 lectin, galactoside- sugar binding, extracellular binding, soluble, 7 space, nucleus, apoptosis, 11. 206400_at LGALS7 (galectin 7) heterophilic cell adhesion <0.01 2 S100 calcium binding calcium ion binding, epidermis 12. 205916_at S100A7 protein A7 (psoriasin 1) development <0.01 S100 calcium binding protein A8 (calgranulin calcium ion binding, extracellular 13. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Download, Or Email Articles for Individual Use
Florida State University Libraries Faculty Publications The Department of Biomedical Sciences 2010 Functional Intersection of the Kallikrein- Related Peptidases (KLKs) and Thrombostasis Axis Michael Blaber, Hyesook Yoon, Maria Juliano, Isobel Scarisbrick, and Sachiko Blaber Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] Article in press - uncorrected proof Biol. Chem., Vol. 391, pp. 311–320, April 2010 • Copyright ᮊ by Walter de Gruyter • Berlin • New York. DOI 10.1515/BC.2010.024 Review Functional intersection of the kallikrein-related peptidases (KLKs) and thrombostasis axis Michael Blaber1,*, Hyesook Yoon1, Maria A. locus (Gan et al., 2000; Harvey et al., 2000; Yousef et al., Juliano2, Isobel A. Scarisbrick3 and Sachiko I. 2000), as well as the adoption of a commonly accepted Blaber1 nomenclature (Lundwall et al., 2006), resolved these two fundamental issues. The vast body of work has associated 1 Department of Biomedical Sciences, Florida State several cancer pathologies with differential regulation or University, Tallahassee, FL 32306-4300, USA expression of individual members of the KLK family, and 2 Department of Biophysics, Escola Paulista de Medicina, has served to elevate the importance of the KLKs in serious Universidade Federal de Sao Paulo, Rua Tres de Maio 100, human disease and their diagnosis (Diamandis et al., 2000; 04044-20 Sao Paulo, Brazil Diamandis and Yousef, 2001; Yousef and Diamandis, 2001, 3 Program for Molecular Neuroscience and Departments of 2003; -

Recombinant Human KLK11/Kallikrein-11 Protein
Leader in Biomolecular Solutions for Life Science Recombinant Human KLK11/Kallikrein-11 Protein Catalog No.: RP00149 Recombinant Sequence Information Background Species Gene ID Swiss Prot kallikrein-related peptidase 11 (KLK11), also known as hippostasin, trypsin-like Human 11012 Q9UBX7 serine protease and PRSS20, is a member of human tissue kallikrein family. It is a subgroup of serine proteases with diverse physiological functions, which is Tags implicated in carcinogenesis and some have potential as novel cancer and other C-6×His disease biomarkers.The KLK11 gene is one of the fifteen kallikrein subfamily members located in a cluster on chromosome 19.Two alternatively spliced forms Synonyms exist, resulting in 250 (isoform 1) and 282 (isoform 2) amino acid PRSS20;TLSP sequences,respectively. Isoform 1 is predominantly expressed in brain whereas isoform 2 is preferentially expressed in prostate. KLK11 is a novel marker for ovarian and prostate cancer carcinomas. Recombinant human KLK11, after being activated by thermolysin, is active against a thioester substrate. This activity can be inhibited by AEBSF dichloroisocoumarin, and aprotinin. Product Information Source Purification Basic Information HEK293 cells > 92% by SDS- PAGE. Description Recombinant Human KLK11/Kallikrein-11 Protein is produced by HEK293 cells Endotoxin expression system. The target protein is expressed with sequence (Met1-Asn250) < 0.1 EU/μg of the protein by LAL of human KLK11/Kallikrein-11 (Accession #NP_006844.1) fused with a 6×His tag at method. the C-terminus. Formulation Bio-Activity Lyophilized from a 0.22 μm filtered solution of 20mM Tris,150mM NaCl, pH Storage 8.0.Contact us for customized product Store the lyophilized protein at -20°C to -80 °C for long term. -
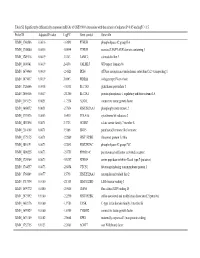
Table SI. Significantly Differentially Expressed Mrnas of GSE19089 Data Series with the Criteria of Adjusted P<0.05 Andlogfc
Table SI. Significantly differentially expressed mRNAs of GSE19089 data series with the criteria of adjusted P<0.05 andlogFC>1.5. Probe ID Adjusted P-value LogFC Gene symbol Gene title ILMN_1740586 0.0016 -3.0593 PTHLH phospholipase A2 group IIA ILMN_1705080 0.0016 -5.0044 PTHLH secreted LY6/PLAUR domain containing 1 ILMN_1749118 0.0019 3.1363 LAMC2 calmodulin like 5 ILMN_1800341 0.0019 2.4138 CALML5 WD repeat domain 66 ILMN_1674460 0.0019 -2.4821 IRX4 ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 ILMN_1675857 0.0019 2.0803 WDR66 collagen type IV 6 chain ILMN_1726666 0.0038 -3.8331 SLC7A8 glutathione peroxidase 3 ILMN_2056606 0.0047 -2.1260 SLC2A1 protein phosphatase 1 regulatory inhibitor subunit 1A ILMN_2115125 0.0051 -1.7534 ALG1L connective tissue growth factor ILMN_1668072 0.0051 -2.7854 HIST2H2AA3 phosphoglycerate mutase 2 ILMN_1739576 0.0053 1.6410 COL4A6 cytochrome b5 reductase 2 ILMN_1807894 0.0071 2.3723 HOXB7 solute carrier family 7 member 8 ILMN_2314169 0.0071 3.5186 ISG15 parathyroid hormone like hormone ILMN_1775175 0.0071 -2.2869 HIST1H2BK ribosomal protein L3 like ILMN_1810191 0.0073 -2.2843 HIST2H2AC phospholipase A2 group IVC ILMN_1800225 0.0073 -2.0751 PPP1R14C peroxisome proliferator activated receptor ILMN_1720984 0.0073 -5.5257 NDRG4 serine peptidase inhibitor Kazal type 7 (putative) ILMN_1764557 0.0073 -2.6854 VTCN1 fat storage inducing transmembrane protein 1 ILMN_1760849 0.0077 1.5798 HIST2H2AA3 neuropilin and tolloid like 2 ILMN_1713534 0.0100 -2.1515 HIST1H2BD LIM domain binding 3 ILMN_1699772 -

Purification and Characterization of Human Kallikrein 11, a Candidate Prostate and Ovarian Cancer Biomarker, from Seminal Plasma Liu-Ying Luo,1, 2 Shannon J.C
Human Cancer Biology Purification and Characterization of Human Kallikrein 11, a Candidate Prostate and Ovarian Cancer Biomarker, from Seminal Plasma Liu-Ying Luo,1, 2 Shannon J.C. Shan,1, 2 Marc B. Elliott,1, 2 Antoninus Soosaipillai,1 and Eleftherios P. Diamandis1, 2 Abstract Purpose: Preliminary data suggest that hK11is a novel serum biomarker for prostate and ovarian cancer. To examine the enzymatic characteristics of hK11, we purified and functionally characterized native hK11from seminal plasma. Experimental Design: hK11was purified from seminal plasma by immunoaffinity chromatogra- phy and characterized by kinetic analysis, electrophoresis,Western blots, and mass spectrometry. Results: hK11is present in seminal plasma at concentrations ranging from 2 to 37 Ag/mL. Using immunoaffinity chromatography and reverse-phase high-performance liquid chromatography, we purified hK11to homogeneity. In seminal plasma, hK11is present as a free enzyme of f40 kDa. About 40% of hK11is enzymatically active, whereas the rest is inactivated by internal cleavage after Arg15 6 (Genbank accession no. AF164623), which generates two peptides of f20 kDa, connected by internal disulfide bonds. Purified hK11possesses trypsin-like activity and cleaves synthetic peptides after arginine but not lysine residues. It does not cleave chymotrypsin substrates. Antithrombin, a1-antichymotrypsin, a2-antiplasmin, and a1-antitrypsin have no effect on hK11activity and do not form complexes with hK11 in vitro. The strongest inhibitor, APMSF, completely inhibited hK11activity at a concentration of 2.5 mmol/L. Aprotinin and an hK11- specific monoclonal antibody inhibited hK11activity up to 40%. Plasmin is a strong candidate for cleaving hK11at Arg15 6. Conclusion: This is the first report on purification and characterization of native hK11.We speculate that hK11,along with other kallikreins, proteases, andinhibitors, participates in a cascade enzymatic pathway responsible for semen liquefaction after ejaculation. -

Activation Profiles and Regulatory Cascades of the Human Kallikrein-Related Peptidases Hyesook Yoon
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2008 Activation Profiles and Regulatory Cascades of the Human Kallikrein-Related Peptidases Hyesook Yoon Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES ACTIVATION PROFILES AND REGULATORY CASCADES OF THE HUMAN KALLIKREIN-RELATED PEPTIDASES By HYESOOK YOON A Dissertation submitted to the Department of Chemistry and Biochemistry in partial fulfillment of the requirements for the degree of Doctor of Philosophy Degree Awarded: Fall Semester, 2008 The members of the Committee approve the dissertation of Hyesook Yoon defended on July 10th, 2008. ________________________ Michael Blaber Professor Directing Dissertation ________________________ Hengli Tang Outside Committee Member ________________________ Brian Miller Committee Member ________________________ Oliver Steinbock Committee Member Approved: ____________________________________________________________ Joseph B. Schlenoff, Chair, Department of Chemistry and Biochemistry The Office of Graduate Studies has verified and approved the above named committee members. ii ACKNOWLEDGMENTS I would like to dedicate this dissertation to my parents for all your support, and my sister and brother. I would also like to give great thank my advisor, Dr. Blaber for his patience, guidance. Without him, I could never make this achievement. I would like to thank to all the members in Blaber lab. They are just like family to me and I deeply appreciate their kindness, consideration and supports. I specially like to thank to Mrs. Sachiko Blaber for her endless guidance and encouragement. I would like to thank Dr Jihun Lee, Margaret Seavy, Rani and Doris Terry for helpful discussions and supports. -

The Usefulness of Serum Human Kallikrein 11 for Discriminating Between Prostate Cancer and Benign Prostatic Hyperplasia
[CANCER RESEARCH 63, 6543–6546, October 1, 2003] The Usefulness of Serum Human Kallikrein 11 for Discriminating between Prostate Cancer and Benign Prostatic Hyperplasia Terukazu Nakamura, Andreas Scorilas, Carsten Stephan, Klaus Jung, Antoninus R. Soosaipillai, and Eleftherios P. Diamandis1 Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Ontario M5G 1X5, Canada [T. N., A. R. S., E. P. D.]; Department of Urology, Kyoto Prefectural University of Medicine, Kyoto, Japan 602-8566 [T. N.]; Department of Biochemistry and Molecular Biology, University of Athens and National Center for Scientific Research “Demokritos,” Athens, Greece 15310 [A. S.]; Department of Urology, University Hospital Charite´, Humboldt University, Berlin, Germany D-10098 [C. S., K. J.]; Department of Laboratory Medicine and Pathobiology, University of Toronto, Ontario M5G IL5, Canada [E. P. D.] ABSTRACT hippostasin (7–9). With the official nomenclature, TLSP/hippostasin is now known as hK11. This protein is encoded by the KLK11 gene, Prostate-specific antigen (PSA) is the most useful tumor marker for which belongs to the human kallikrein family along with PSA (hK3) diagnosis and monitoring of prostate cancer (CaP). Recently, we devel- and other kallikreins (10). We have previously demonstrated that oped a specific immunoassay for human kallikrein 11 (hK11), one of the kallikrein gene family members, and found that hK11 was highly ex- hK11 protein is highly expressed in the prostate (8, 11). We have also pressed in prostatic tissue and could be detected in seminal plasma (E. P. developed an hK11-specific immunoassay and quantified hK11 in Diamandis et al., Cancer Res., 62: 295–300, 2002). The aim of this study seminal plasma and prostatic tissue extracts.