Cloud-Clone Productlist Viroquest Inc
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

An Overview of the Kallikrein Gene Families in Humans and Other Species: Emerging Candidate Tumour Markers૾
Clinical Biochemistry 36 (2003) 443–452 An overview of the kallikrein gene families in humans and other species: Emerging candidate tumour markers૾ George M. Yousefa,b, Eleftherios P. Diamandisa,b,* aDepartment of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Ontario, Canada bDepartment of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Ontario, Canada Abstract Kallikreins are serine proteases with diverse physiologic functions. They are represented by multigene families in many animal species, especially in rat and mouse. Recently, the human kallikrein gene family has been fully characterized and includes 15 members, tandemly localized on chromosome 19q13.4. A new definition has now been proposed for kallikreins, which is not based on function but, rather, on close proximity and structural similarities. In this review, we summarize available information about kallikreins in many animal species with special emphasis on human kallikreins. We discuss the common structural features of kallikreins at the DNA, mRNA and protein levels and overview their evolutionary history. Kallikreins are expressed in a wide range of tissues including the salivary gland, endocrine or endocrine-related tissues such as testis, prostate, breast and endometrium and in the central nervous system. Most, if not all, genes are under steroid hormone regulation. Accumulating evidence indicates that kallikreins are involved in many pathologic conditions. Of special interest is the potential role of kallikreins in the central nervous system. In addition, many kallikreins seem to be candidate tumor markers for many malignancies, especially those of endocrine-related organs. © 2003 The Canadian Society of Clinical Chemists. All rights reserved. Keywords: Kallikrein; Tumor markers; Cancer biomarkers; Prostate cancer; Breast cancer; Ovarian cancer; Alzheimer’s disease; Serine proteases; Chromosome 19; Kallikrein evolution; Rodent kallikreins; Hormonally regulated genes 1. -
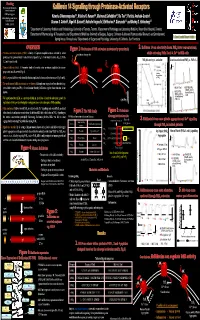
AACR2006-1686P
Funding Proteinases and Inflammation Network Group grant Kallikrein 14 Signalling through Proteinase-Activated Receptors CIHR operating grant 1,2 3,4 3,4 3,4 5 Alberta Heritage Foundation for Katerina Oikonomopoulou , Kristina K. Hansen , Mahmoud Saifeddine , Illa Tea , Patricia Andrade-Gordon , Medical Research Graeme S. Cottrell6, Nigel W. Bunnett6, Nathalie Vergnolle3, Eleftherios P. Diamandis1,2 and Morley D. Hollenberg3,4 NSERC / CRSNG 1Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto; 2Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto; 3Department of Pharmacology & Therapeutics, and 4Department of Medicine, University of Calgary, Calgary; 5Johnson & Johnson Pharmaceutical Research and Development, Spring House, Pennsylvania, 6Departments of Surgery and Physiology, University of California, San Francisco Kallikrein 14 can selectively disarm PAR (lower concentrations), OVERVIEW Figure 1: Mechanism of PAR activation (activation by proteolysis) 2. Kallikrein 14 can selectively disarm PAR1 (lower concentrations), 2+ Proteinase-activated receptors (PARs): a family of G-protein coupled receptors activated by serine proteinase cleavage site whilst activating PARs 2 and 4; Ca in HEK cells proteinases via a proteolytically revealed ‘tethered ligand’ (Fig. 1). Four family members (Fig. 2); PARs N PAR disarming 1, 2 and 4 signal to cells. N PAR1 dis-arming1 vs. activation Selective activation of PAR4 vs. PARs 1, 2 Human kallikreins (hKs): A 15-member family of secreted serine proteinases implicated in tumour 1 cm Thrombin 1 cm 2 min 2 min TFLLR-NH2 progression and cell survival (Fig. 4). (selective desensitization hK14: a tryptic kallikrein; wide tissue distribution, implicated in breast and ovarian cancer (Fig. 5 and 6). of PAR1) The mechanism of kallikrein action is not yet known: Although some targets have been identified (e.g. -

Whole Exome Sequencing in Families at High Risk for Hodgkin Lymphoma: Identification of a Predisposing Mutation in the KDR Gene
Hodgkin Lymphoma SUPPLEMENTARY APPENDIX Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene Melissa Rotunno, 1 Mary L. McMaster, 1 Joseph Boland, 2 Sara Bass, 2 Xijun Zhang, 2 Laurie Burdett, 2 Belynda Hicks, 2 Sarangan Ravichandran, 3 Brian T. Luke, 3 Meredith Yeager, 2 Laura Fontaine, 4 Paula L. Hyland, 1 Alisa M. Goldstein, 1 NCI DCEG Cancer Sequencing Working Group, NCI DCEG Cancer Genomics Research Laboratory, Stephen J. Chanock, 5 Neil E. Caporaso, 1 Margaret A. Tucker, 6 and Lynn R. Goldin 1 1Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 2Cancer Genomics Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 3Ad - vanced Biomedical Computing Center, Leidos Biomedical Research Inc.; Frederick National Laboratory for Cancer Research, Frederick, MD; 4Westat, Inc., Rockville MD; 5Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; and 6Human Genetics Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA ©2016 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2015.135475 Received: August 19, 2015. Accepted: January 7, 2016. Pre-published: June 13, 2016. Correspondence: [email protected] Supplemental Author Information: NCI DCEG Cancer Sequencing Working Group: Mark H. Greene, Allan Hildesheim, Nan Hu, Maria Theresa Landi, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Anand Pathak, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, Xiaohong R. Yang, Guoqin Yu NCI DCEG Cancer Genomics Research Laboratory: Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Download, Or Email Articles for Individual Use
Florida State University Libraries Faculty Publications The Department of Biomedical Sciences 2010 Functional Intersection of the Kallikrein- Related Peptidases (KLKs) and Thrombostasis Axis Michael Blaber, Hyesook Yoon, Maria Juliano, Isobel Scarisbrick, and Sachiko Blaber Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] Article in press - uncorrected proof Biol. Chem., Vol. 391, pp. 311–320, April 2010 • Copyright ᮊ by Walter de Gruyter • Berlin • New York. DOI 10.1515/BC.2010.024 Review Functional intersection of the kallikrein-related peptidases (KLKs) and thrombostasis axis Michael Blaber1,*, Hyesook Yoon1, Maria A. locus (Gan et al., 2000; Harvey et al., 2000; Yousef et al., Juliano2, Isobel A. Scarisbrick3 and Sachiko I. 2000), as well as the adoption of a commonly accepted Blaber1 nomenclature (Lundwall et al., 2006), resolved these two fundamental issues. The vast body of work has associated 1 Department of Biomedical Sciences, Florida State several cancer pathologies with differential regulation or University, Tallahassee, FL 32306-4300, USA expression of individual members of the KLK family, and 2 Department of Biophysics, Escola Paulista de Medicina, has served to elevate the importance of the KLKs in serious Universidade Federal de Sao Paulo, Rua Tres de Maio 100, human disease and their diagnosis (Diamandis et al., 2000; 04044-20 Sao Paulo, Brazil Diamandis and Yousef, 2001; Yousef and Diamandis, 2001, 3 Program for Molecular Neuroscience and Departments of 2003; -

C:\Users\Administrator\Desktop\Array Datasheet\Disease Gene Array
Product Data Sheet ExProfileTM Human Proteases Related Gene qPCR Array For focused group profiling of human proteases genes expression Cat. No. QG096-A (6 x 96-well plate, Format A) Cat. No. QG096-B (6 x 96-well plate, Format B) Cat. No. QG096-C (6 x 96-well plate, Format C) Cat. No. QG096-D (6 x 96-well plate, Format D) Cat. No. QG096-E (6 x 96-well plate, Format E) Plates available individually or as a set of 6. Each set contains 504 unique gene primer pairs deposited in one 96-well plate. Introduction The ExProfile human proteases related gene qPCR array profiles the expression of 504 human genes related to proteases. These genes are carefully chosen for their close correlation based on a thorough literature search of peer-reviewed publications, mainly including genes that encode various proteases. This array allows researchers to study the related genes to gain understanding of their roles in the functioning and characterization of proteases. QG096 plate 01: 84 unique gene PCR primer pairs QG096 plate 02: 84 unique gene PCR primer pairs QG096 plate 03: 84 unique gene PCR primer pairs QG096 plate 04: 84 unique gene PCR primer pairs QG096 plate 05: 84 unique gene PCR primer pairs QG096 plate 06: 84 unique gene PCR primer pairs Shipping and storage condition Shipped at room temperate Stable for at least 6 months when stored at -20°C Array format GeneCopoeia provides five qPCR array formats (A, B, C, D, and E) suitable for use with the following real- time cyclers. -
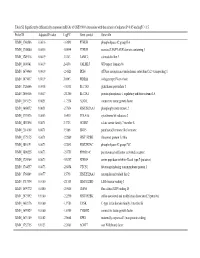
Table SI. Significantly Differentially Expressed Mrnas of GSE19089 Data Series with the Criteria of Adjusted P<0.05 Andlogfc
Table SI. Significantly differentially expressed mRNAs of GSE19089 data series with the criteria of adjusted P<0.05 andlogFC>1.5. Probe ID Adjusted P-value LogFC Gene symbol Gene title ILMN_1740586 0.0016 -3.0593 PTHLH phospholipase A2 group IIA ILMN_1705080 0.0016 -5.0044 PTHLH secreted LY6/PLAUR domain containing 1 ILMN_1749118 0.0019 3.1363 LAMC2 calmodulin like 5 ILMN_1800341 0.0019 2.4138 CALML5 WD repeat domain 66 ILMN_1674460 0.0019 -2.4821 IRX4 ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 ILMN_1675857 0.0019 2.0803 WDR66 collagen type IV 6 chain ILMN_1726666 0.0038 -3.8331 SLC7A8 glutathione peroxidase 3 ILMN_2056606 0.0047 -2.1260 SLC2A1 protein phosphatase 1 regulatory inhibitor subunit 1A ILMN_2115125 0.0051 -1.7534 ALG1L connective tissue growth factor ILMN_1668072 0.0051 -2.7854 HIST2H2AA3 phosphoglycerate mutase 2 ILMN_1739576 0.0053 1.6410 COL4A6 cytochrome b5 reductase 2 ILMN_1807894 0.0071 2.3723 HOXB7 solute carrier family 7 member 8 ILMN_2314169 0.0071 3.5186 ISG15 parathyroid hormone like hormone ILMN_1775175 0.0071 -2.2869 HIST1H2BK ribosomal protein L3 like ILMN_1810191 0.0073 -2.2843 HIST2H2AC phospholipase A2 group IVC ILMN_1800225 0.0073 -2.0751 PPP1R14C peroxisome proliferator activated receptor ILMN_1720984 0.0073 -5.5257 NDRG4 serine peptidase inhibitor Kazal type 7 (putative) ILMN_1764557 0.0073 -2.6854 VTCN1 fat storage inducing transmembrane protein 1 ILMN_1760849 0.0077 1.5798 HIST2H2AA3 neuropilin and tolloid like 2 ILMN_1713534 0.0100 -2.1515 HIST1H2BD LIM domain binding 3 ILMN_1699772 -

Activation Profiles and Regulatory Cascades of the Human Kallikrein-Related Peptidases Hyesook Yoon
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2008 Activation Profiles and Regulatory Cascades of the Human Kallikrein-Related Peptidases Hyesook Yoon Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES ACTIVATION PROFILES AND REGULATORY CASCADES OF THE HUMAN KALLIKREIN-RELATED PEPTIDASES By HYESOOK YOON A Dissertation submitted to the Department of Chemistry and Biochemistry in partial fulfillment of the requirements for the degree of Doctor of Philosophy Degree Awarded: Fall Semester, 2008 The members of the Committee approve the dissertation of Hyesook Yoon defended on July 10th, 2008. ________________________ Michael Blaber Professor Directing Dissertation ________________________ Hengli Tang Outside Committee Member ________________________ Brian Miller Committee Member ________________________ Oliver Steinbock Committee Member Approved: ____________________________________________________________ Joseph B. Schlenoff, Chair, Department of Chemistry and Biochemistry The Office of Graduate Studies has verified and approved the above named committee members. ii ACKNOWLEDGMENTS I would like to dedicate this dissertation to my parents for all your support, and my sister and brother. I would also like to give great thank my advisor, Dr. Blaber for his patience, guidance. Without him, I could never make this achievement. I would like to thank to all the members in Blaber lab. They are just like family to me and I deeply appreciate their kindness, consideration and supports. I specially like to thank to Mrs. Sachiko Blaber for her endless guidance and encouragement. I would like to thank Dr Jihun Lee, Margaret Seavy, Rani and Doris Terry for helpful discussions and supports. -

A Multiparametric Serum Kallikrein Panel for Diagnosis of Non ^ Small
Imaging, Diagnosis, Prognosis A Multiparametric Serum Kallikrein Panel for Diagnosis of Non ^ Small Cell Lung Carcinoma Chris Planque,1, 2 Lin Li,3 Yingye Zheng,3 Antoninus Soosaipillai,1, 2 Karen Reckamp,4 David Chia,5 Eleftherios P. Diamandis,1, 2 and Lee Goodglick5 Abstract Purpose: Human tissue kallikreins are a family of15 secreted serine proteases.We have previous- ly shown that the expression of several tissue kallikreins is significantly altered at the transcription- al level in lung cancer. Here, we examined the clinical value of 11members of the tissue kallikrein family as potential biomarkers for lung cancer diagnosis. Experimental Design: Serum specimens from 51 patients with non ^ small cell lung cancer (NSCLC) and from 50 healthy volunteers were collected. Samples were analyzed for11kallikreins (KLK1, KLK4-8, and KLK10-14) by specific ELISA. Data were statistically compared and receiver operating characteristic curves were constructed for each kallikrein and for various combinations. Results: Compared with sera from normal subjects, sera of patients with NSCLC had lower levels of KLK5, KLK7, KLK8, KLK10, and KLK12, and higher levels of KLK11, KLK13, and KLK14. Expres- sion of KLK11and KLK12 was positively correlated with stage.With the exception of KLK5, expres- sion of kallikreins was independent of smoking status and gender. KLK11, KLK12, KLK13, and KLK14 were associated with higher risk of NSCLC as determined by univariate analysis and con- firmed by multivariate analysis.The receiver operating characteristic curve of KLK4, KLK8, KLK10, KLK11,KLK12, KLK13, and KLK14 combined exhibited an area under the curve of 0.90 (95% con- fidence interval, 0.87-0.97). -

A Genomic Analysis of Rat Proteases and Protease Inhibitors
A genomic analysis of rat proteases and protease inhibitors Xose S. Puente and Carlos López-Otín Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, 33006-Oviedo, Spain Send correspondence to: Carlos López-Otín Departamento de Bioquímica y Biología Molecular Facultad de Medicina, Universidad de Oviedo 33006 Oviedo-SPAIN Tel. 34-985-104201; Fax: 34-985-103564 E-mail: [email protected] Proteases perform fundamental roles in multiple biological processes and are associated with a growing number of pathological conditions that involve abnormal or deficient functions of these enzymes. The availability of the rat genome sequence has opened the possibility to perform a global analysis of the complete protease repertoire or degradome of this model organism. The rat degradome consists of at least 626 proteases and homologs, which are distributed into five catalytic classes: 24 aspartic, 160 cysteine, 192 metallo, 221 serine, and 29 threonine proteases. Overall, this distribution is similar to that of the mouse degradome, but significatively more complex than that corresponding to the human degradome composed of 561 proteases and homologs. This increased complexity of the rat protease complement mainly derives from the expansion of several gene families including placental cathepsins, testases, kallikreins and hematopoietic serine proteases, involved in reproductive or immunological functions. These protease families have also evolved differently in the rat and mouse genomes and may contribute to explain some functional differences between these two closely related species. Likewise, genomic analysis of rat protease inhibitors has shown some differences with the mouse protease inhibitor complement and the marked expansion of families of cysteine and serine protease inhibitors in rat and mouse with respect to human. -

Prognostic Significance of Multiple Kallikreins in High-Grade Astrocytoma Kristen L
Drucker et al. BMC Cancer (2015) 15:565 DOI 10.1186/s12885-015-1566-5 RESEARCH ARTICLE Open Access Prognostic significance of multiple kallikreins in high-grade astrocytoma Kristen L. Drucker1, Caterina Gianinni2, Paul A. Decker3, Eleftherios P. Diamandis4 and Isobel A. Scarisbrick1,5* Abstract Background: Kallikreins have clinical value as prognostic markers in a subset of malignancies examined to date, including kallikrein 3 (prostate specific antigen) in prostate cancer. We previously demonstrated that kallikrein 6 is expressed at higher levels in grade IV compared to grade III astrocytoma and is associated with reduced survival of GBM patients. Methods: In this study we determined KLK1, KLK6, KLK7, KLK8, KLK9 and KLK10 protein expression in two independent tissue microarrays containing 60 grade IV and 8 grade III astrocytoma samples. Scores for staining intensity, percent of tumor stained and immunoreactivity scores (IR, product of intensity and percent) were determined and analyzed for correlation with patient survival. Results: Grade IV glioma was associated with higher levels of kallikrein-immunostaining compared to grade III specimens. Univariable Cox proportional hazards regression analysis demonstrated that elevated KLK6- or KLK7-IR was associated with poor patient prognosis. In addition, an increased percent of tumor immunoreactive for KLK6 or KLK9 was associated with decreased survival in grade IV patients. Kaplan-Meier survival analysis indicated that patients with KLK6-IR < 10, KLK6 percent tumor core stained < 3, or KLK7-IR < 9 had a significantly improved survival. Multivariable analysis indicated that the significance of these parameters was maintained even after adjusting for gender and performance score. Conclusions: These data suggest that elevations in glioblastoma KLK6, KLK7 and KLK9 protein have utility as prognostic markers of patient survival.