The Open Access Israeli Journal of Aquaculture – Bamidgeh
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

First Report of the Non-Native Midas Cichlid, Amphilophus Citrinellus (Gunther, 1864), in Laguna De Bay, Philippines
First Report of the Non-native Midas Cichlid, Amphilophus citrinellus (Gunther, 1864), in Laguna de Bay, Philippines Item Type article Authors Poniente, Jennifer A.; Dela Peña, John T.; Pol, Romualdo M.; Zapanta, Levita H.; Santos, Mudjekeewis D. DOI 10.31398/tpjf/26.2.2019A0007 Download date 04/10/2021 12:53:05 Link to Item http://hdl.handle.net/1834/41171 The Philippine Journal of Fisheries 26(2): 55-60 July - December 2019 DOI: 10.31398/tpjf/26.2.2019A0007 SHORT COMMUNICATION First report of the Non-native Midas Cichlid, Amphilophus citrinellus (Gunther, 1864), in Laguna de Bay, Philippines Jennifer A. Poniente1, John T. Dela Peña1, Romualdo M. Pol2, Levita H. Zapanta2+ and Mudjekeewis D. Santos1* 1Genetic Fingerprinting Laboratory, National Fisheries Research and Development Institute, Quezon City, Philippines 2National Inland Fisheries Technology Center, Bureau of Fisheries and Aquatic Resources, Tanay, Rizal, Philippines ABSTRACT In recent years, increasing global trade, travel, and transport had rapidly increased the rate of introduction and diversity of non-native fish species. Once established, some introduced fish species can become aggressive and dangerously invasive. Here, we provide the first report of the occurrence of a non-native Midas cichlid Amphilophus citrinellus in Laguna de Bay using morphological analysis and genetic marker, specifically the mitochondrial gene cytochrome c oxidase subunit 1 (CO1). The results provide important information on the presence of another invasive species in Laguna de Bay that needs to be addressed since this species can competitively exclude, predate, and displace native species. E-mail address: [email protected]* Keywords: Amphilophus citrinellus, non-native fish, Received: October 3, 2019 Laguna de Bay, invasive species, cichlid Accepted: December 17, 2019 on-native species may cause changes in the (1976) reported that the natural food supply of the ecosystems to which they are introduced lake could sustain the rearing of milkfish in the lake. -

Field Guide to the Nonindigenous Marine Fishes of Florida
Field Guide to the Nonindigenous Marine Fishes of Florida Schofield, P. J., J. A. Morris, Jr. and L. Akins Mention of trade names or commercial products does not constitute endorsement or recommendation for their use by the United States goverment. Pamela J. Schofield, Ph.D. U.S. Geological Survey Florida Integrated Science Center 7920 NW 71st Street Gainesville, FL 32653 [email protected] James A. Morris, Jr., Ph.D. National Oceanic and Atmospheric Administration National Ocean Service National Centers for Coastal Ocean Science Center for Coastal Fisheries and Habitat Research 101 Pivers Island Road Beaufort, NC 28516 [email protected] Lad Akins Reef Environmental Education Foundation (REEF) 98300 Overseas Highway Key Largo, FL 33037 [email protected] Suggested Citation: Schofield, P. J., J. A. Morris, Jr. and L. Akins. 2009. Field Guide to Nonindigenous Marine Fishes of Florida. NOAA Technical Memorandum NOS NCCOS 92. Field Guide to Nonindigenous Marine Fishes of Florida Pamela J. Schofield, Ph.D. James A. Morris, Jr., Ph.D. Lad Akins NOAA, National Ocean Service National Centers for Coastal Ocean Science NOAA Technical Memorandum NOS NCCOS 92. September 2009 United States Department of National Oceanic and National Ocean Service Commerce Atmospheric Administration Gary F. Locke Jane Lubchenco John H. Dunnigan Secretary Administrator Assistant Administrator Table of Contents Introduction ................................................................................................ i Methods .....................................................................................................ii -

A Practical Handbook for Determining the Ages of Gulf of Mexico And
A Practical Handbook for Determining the Ages of Gulf of Mexico and Atlantic Coast Fishes THIRD EDITION GSMFC No. 300 NOVEMBER 2020 i Gulf States Marine Fisheries Commission Commissioners and Proxies ALABAMA Senator R.L. “Bret” Allain, II Chris Blankenship, Commissioner State Senator District 21 Alabama Department of Conservation Franklin, Louisiana and Natural Resources John Roussel Montgomery, Alabama Zachary, Louisiana Representative Chris Pringle Mobile, Alabama MISSISSIPPI Chris Nelson Joe Spraggins, Executive Director Bon Secour Fisheries, Inc. Mississippi Department of Marine Bon Secour, Alabama Resources Biloxi, Mississippi FLORIDA Read Hendon Eric Sutton, Executive Director USM/Gulf Coast Research Laboratory Florida Fish and Wildlife Ocean Springs, Mississippi Conservation Commission Tallahassee, Florida TEXAS Representative Jay Trumbull Carter Smith, Executive Director Tallahassee, Florida Texas Parks and Wildlife Department Austin, Texas LOUISIANA Doug Boyd Jack Montoucet, Secretary Boerne, Texas Louisiana Department of Wildlife and Fisheries Baton Rouge, Louisiana GSMFC Staff ASMFC Staff Mr. David M. Donaldson Mr. Bob Beal Executive Director Executive Director Mr. Steven J. VanderKooy Mr. Jeffrey Kipp IJF Program Coordinator Stock Assessment Scientist Ms. Debora McIntyre Dr. Kristen Anstead IJF Staff Assistant Fisheries Scientist ii A Practical Handbook for Determining the Ages of Gulf of Mexico and Atlantic Coast Fishes Third Edition Edited by Steve VanderKooy Jessica Carroll Scott Elzey Jessica Gilmore Jeffrey Kipp Gulf States Marine Fisheries Commission 2404 Government St Ocean Springs, MS 39564 and Atlantic States Marine Fisheries Commission 1050 N. Highland Street Suite 200 A-N Arlington, VA 22201 Publication Number 300 November 2020 A publication of the Gulf States Marine Fisheries Commission pursuant to National Oceanic and Atmospheric Administration Award Number NA15NMF4070076 and NA15NMF4720399. -

Oreochromis Rukwaensis) Ecological Risk Screening Summary
Lake Rukwa Tilapia (Oreochromis rukwaensis) Ecological Risk Screening Summary U.S. Fish & Wildlife Service, March 2012 Revised, July 2018 Web Version, 6/4/2020 Organism Type: Fish Overall Risk Assessment Category: Uncertain 1 Native Range and Status in the United States Native Range From Froese and Pauly (2018): “Africa: Lake Rukwa in Tanzania.” From Shechonge et al. (2019): “Oreochromis rukwaensis (Hilgendorf & Pappenheim 1903) previously known only from Lake Rukwa was present in an upstream section of the Ruaha river system, where a major exploited population was recorded at the Mtera Dam Lake [Tanzania].” Status in the United States No records of Oreochromis rukwaensis occurrences in the United States were found. No information on trade of O. rukwaensis in the United States was found. 1 The Florida Fish and Wildlife Conservation Commission has listed the tilapia, Oreochromis rukwaensis as a prohibited species. Prohibited nonnative species (FFWCC 2020), "are considered to be dangerous to the ecology and/or the health and welfare of the people of Florida. These species are not allowed to be personally possessed or used for commercial activities." Means of Introductions in the United States No records of Oreochromis rukwaensis occurrences in the United States were found. Remarks No additional remarks. 2 Biology and Ecology Taxonomic Hierarchy and Taxonomic Standing According to Eschmeyer et al. (2018), Oreochromis rukwaensis (Hilgendorf and Pappenheim 1903) is the current valid name of this species. From ITIS (2018): Kingdom Animalia -

Lake Chala Tilapia (Oreochromis Hunteri) Ecological Risk Screening Summary
Lake Chala Tilapia (Oreochromis hunteri) Ecological Risk Screening Summary U.S. Fish & Wildlife Service, March 2012 Revised, June 2018 Web Version, 12/15/2020 Organism Type: Fish Overall Risk Assessment Category: Uncertain Photo: D. H. Eccles. Licensed under Creative Commons BY-NC 3.0. Available: http://www.fishbase.org/photos/PicturesSummary.php?StartRow=0&ID=2032&what=species&T otRec=2. (June 18, 2018). 1 Native Range and Status in the United States Native Range From Froese and Pauly (2018a): “Africa: endemic to Lake Chala [Seegers et al. 2003].” 1 Status in the United States No records of Oreochromis hunteri in trade or in the wild in the United States were found. The Florida Fish and Wildlife Conservation Commission has listed the tilapia Oreochromis hunteri as a prohibited species. Prohibited nonnative species (FFWCC 2018), "are considered to be dangerous to the ecology and/or the health and welfare of the people of Florida. These species are not allowed to be personally possessed or used for commercial activities. All species in the genus Oreochromis are considered regulated Type A species in Washington. Regulated Type A species (Washington State Senate 2019) are “nonnative aquatic animal species that pose a low to moderate invasive risk that can be managed based on intended use or geographic scope of introduction, have a beneficial use, and are a priority for department-led or department-approved management of the species' beneficial use and invasive risks.” Possession of any species of tilapia is prohibited without permit in the State of Louisiana (Louisiana State Legislature 2019). O. amphimelas falls within Group I of New Mexico’s Department of Game and Fish Director’s Species Importation List (New Mexico Department of Game and Fish 2010). -

A BIBLIOGRAPHY of IMPORTANT TILAPIAS (PISCES: CICHLIDAE) for AQUACULTURE Oreochromisvariabilis, 0 Andersoni, 0
AMV'__ BIBLIOGRAPHIES 6 A BIBLIOGRAPHY OF IMPORTANT TILAPIAS (PISCES: CICHLIDAE) FOR AQUACULTURE Oreochromisvariabilis, 0 andersoni, 0. esculentus, 0. leucostictus, 0. rortimer, 0. spilurus niger,Sarotherodon melanotheron and Tilapia sparnmani PETER SCHOENEN INTERNATIONAL CENTER FOR LIVING AQUATIC RESOURCES MANAGEMENT A BIBLIOGRAPHY OF IMPORTANT TILAPIAS (PISCES: CICHLIDAE) FOR AQUACULTURE Oreochromls variabilis, 0. andersoni, 0. esculentus, 0. leucostictus, 0. mortimeri, 0. spilurus niger, Saro therodon melano theron and Tilapia sparrmanii Peter Schoenen International Collection "Cichlid Papers" The Referencc Service Parkstr. 15 D-5176 Inden 4 Federal Republic of Germany 1985 INTERNATIONAL CENTER FOR LIVING AQUATIC RESOURCES MANAGEMENT MANILA, PHILIPPINES A bibliography of important tilapias (Pisces: Cichlidae) for aquaculture Oreochromis variabilis, 0. andersonii, 0. esculentus, 0. leucostictus, 0. mort/tmer, 0. spilunis niger, Sarotherodon melanothero,, ard -/ilapiasparrmanii PETER SCHOENEN Published by the International Center for Living Aquatic Resources Management, MCC P.O. Box 1501, Makati, Metro Manila, Philippines with financial assistance from the International Development Research Centre of Canada through ICLARM's Selective Information Service project. 1985 Printed in Manila, Philippins This bibliography is produced directly from the author's manuscript in oider to provide tilapia workers with a useful document in the shortest time. The author should be consulted in the event of difficulty ir verifying details of particular references or in locating sources. ISSN 0115-5997 ISBN 971-1022-19-2 Schoenen, P. 1985, A bibliography of important tilapias (Pisces: Cichlidae) for aquaculture Oreochromis variabilis, 0. andersonii, 0. esculentus, 0. leucostictus, 0. mortimeri, 0. spilurut niger, Sarotherodon mela. notheron and Tilapia sparrrnanii. ICLAHM Biblio graphies 6,99 p. International Center for Living Aquatic Resources Management, Manila, Philippines. -

Implications for Management AFRICAN GREAT LAKES
AFRICAN GREAT LAKES CONFERENCE 2nd – 5th MAY 2017, ENTEBBE, UGANDA Dynamics of Fish Stocks of Commercial Importance in Lake Victoria, East Africa: Implications for Management Robert Kayanda, Anton Taabu-Munyaho, Dismas Mbabazi, Hillary Mrosso, and Chrisphine Nyamweya INTRODUCTION • Lake Victoria with a surface area of 68,800 sqkm is the world’s second largest freshwater body • It supports one of the world’s most productive inland fisheries with the estimated total fish landings from the lake for the period of 2011 to 2014 have been about 1 million tons with a beach value increasing from about US$ 550 Million in 2011 to about US$ 840 million in 2014. • It supports about 220,000 fishers (Frame Survey 2016) • The fish stocks of Lake Victoria have changed dramatically since the introduction of Nile perch Lates niloticus during the late 1950s and early 1960s Fishery Haplochromines The Original Fish Fauna Brycinus sp Protopterus Rastrineobola Mormyrus spp Barbus spp Bagrus docmac Labeo Schilbe intermedius Oreochromis variabilis Clarias gariepinus Mormyrus spp Synodontis victoriae Oreochromis leucostictus INTRODUCTION Currently, the fisheries is dominated by four major commercial important species, these are; •Nile perch •Dagaa •Nile tilapia •Haplochromis Apart from Nile tilapia only estimated through trawl and catch surveys, the other 3 are estimated through trawl, acoustics, and catch INTRODUCTION This paper summarizes current knowledge of the status of the fish stocks and reviews the need for species specific management plans for the major commercial important fish species of Lake Victoria (Nile perch, Nile tilapia, dagaa and haplochromines). Methods • Fisheries dependent – Frame surveys – Catch assessment surveys • Fisheries independent – Acoustic – Bottom trawl Biomass and relative abundance • Total biomass from the surveys 3500 remained fairly stable over time. -

Fish, Various Invertebrates
Zambezi Basin Wetlands Volume II : Chapters 7 - 11 - Contents i Back to links page CONTENTS VOLUME II Technical Reviews Page CHAPTER 7 : FRESHWATER FISHES .............................. 393 7.1 Introduction .................................................................... 393 7.2 The origin and zoogeography of Zambezian fishes ....... 393 7.3 Ichthyological regions of the Zambezi .......................... 404 7.4 Threats to biodiversity ................................................... 416 7.5 Wetlands of special interest .......................................... 432 7.6 Conservation and future directions ............................... 440 7.7 References ..................................................................... 443 TABLE 7.2: The fishes of the Zambezi River system .............. 449 APPENDIX 7.1 : Zambezi Delta Survey .................................. 461 CHAPTER 8 : FRESHWATER MOLLUSCS ................... 487 8.1 Introduction ................................................................. 487 8.2 Literature review ......................................................... 488 8.3 The Zambezi River basin ............................................ 489 8.4 The Molluscan fauna .................................................. 491 8.5 Biogeography ............................................................... 508 8.6 Biomphalaria, Bulinis and Schistosomiasis ................ 515 8.7 Conservation ................................................................ 516 8.8 Further investigations ................................................. -

Trophic Niche Segregation in the Nilotic Ichthyofauna of Lake Albert (Uganda, Africa)
Environmental Biology of Fishes (2005) 74:247–260 Ó Springer 2005 DOI 10.1007/s10641-005-3190-8 Trophic niche segregation in the Nilotic ichthyofauna of Lake Albert (Uganda, Africa) Linda M. Campbella,d, Sylvester B. Wanderab, Robert J. Thackerc,e, D. George Dixona & Robert E. Heckya aDepartment of Biology, University of Waterloo, 200 University Avenue. Waterloo, Ontario, Canada N2L 3G1 bFisheries Resources Research Institute, P.O. Box 343, Jinja, Uganda cDepartment of Physics and Astronomy, McMaster University, 1280 Main St. W, Hamilton, Ontario, Canada dCurrent address: School of Environmental Studies and Department of Biology, Queen’s University, Kingston, ON, Canada K7L 3N6 (e-mail: [email protected]) eCurrent address: Department of Physics and Astronomy, Queen’s University, Kingston, ON, Canada K7L 3N6 Received 29 April 2004 Accepted 13 February 2005 Key words: d13C, d15N, food webs, Nile perch, stable isotopes Synopsis Nile perch, Lates niloticus, and Nile tilapia, Oreochromis niloticus, were originally transplanted from Lake Albert in western Uganda to the African Great Lakes, Lake Victoria and Lake Kyoga, where they are partially implicated in reduction of the fish species diversity. Lake Albert is facing multiple environmental changes, including declining fish species diversity, hyper-eutrophication, hypoxia, and reduced fish catches. To examine the role of Nile perch and Nile tilapia in the food web in their native Lake Albert, we estimated their diets using stable nitrogen and carbon isotopes. In Lake Albert, the tilapiine congeners (closely related species), Tilapia zillii, Oreochromis leucostictus, and Sarethorodon galilaeus, and the centropomid Nile perch congener, Lates macrophthalmus, have narrower diet breath in the presence of the native O. -
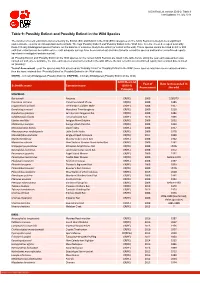
Table 9: Possibly Extinct and Possibly Extinct in the Wild Species
IUCN Red List version 2019-2: Table 9 Last Updated: 18 July 2019 Table 9: Possibly Extinct and Possibly Extinct in the Wild Species The number of recent extinctions documented by the Extinct (EX) and Extinct in the Wild (EW) categories on The IUCN Red List is likely to be a significant underestimate, even for well-known taxa such as birds. The tags 'Possibly Extinct' and 'Possibly Extinct in the Wild' have therefore been developed to identify those Critically Endangered species that are, on the balance of evidence, likely to be extinct (or extinct in the wild). These species cannot be listed as EX or EW until their extinction can be confirmed (i.e., until adequate surveys have been carried out and have failed to record the species and local or unconfirmed reports have been investigated and discounted). All 'Possibly Extinct' and 'Possibly Extinct in the Wild' species on the current IUCN Red List are listed in the table below, along the year each assessment was carried out and, where available, the date each species was last recorded in the wild. Where the last record is an unconfirmed report, last recorded date is noted as "possibly". Year of Assessment - year the species was first assessed as 'Possibly Extinct' or 'Possibly Extinct in the Wild'; some species may have been reassessed since then but have retained their 'Possibly Extinct' or 'Possibly Extinct in the Wild' status. CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild), IUCN Red List Year of Date last recorded in -

Incorporating Biodiversity Concerns in Fisheries Management
Efforts to incorporate Biodiversity Concerns in Management of the Fisheries of Lake Victoria, East Africa Richard Ogutu-Ohwayo National Agricultural Research Organization, Fisheries Resources Research Institute, P. O. Box 343, Jinja, Uganda. E-mail: [email protected] Table of Contents Table of Contents...................................................................................................................................... 1 Abstract ..................................................................................................................................................... 3 Relationships between Components of Aquatic Systems ...................................................................... 3 Geographic Setting of Lake Victoria....................................................................................................... 3 Importance of Fisheries and Biodiversity in Fisheries ...........................................................................4 The Early Fisheries, their Exploitation and Management ...................................................................... 4 Status and Past Trends due to Human Exploitation and Management Efforts for Lake Victoria..... 5 Trends in Non-target Biodiversity Concerns .......................................................................................... 7 The Original Institutional, Policy and Legal Framework....................................................................... 8 Lessons from Past Efforts to Manage the Fisheries of Lake Victoria................................................... -

Out of Lake Tanganyika: Endemic Lake Fishes Inhabit Rapids of the Lukuga River
355 Ichthyol. Explor. Freshwaters, Vol. 22, No. 4, pp. 355-376, 5 figs., 3 tabs., December 2011 © 2011 by Verlag Dr. Friedrich Pfeil, München, Germany – ISSN 0936-9902 Out of Lake Tanganyika: endemic lake fishes inhabit rapids of the Lukuga River Sven O. Kullander* and Tyson R. Roberts** The Lukuga River is a large permanent river intermittently serving as the only effluent of Lake Tanganyika. For at least the first one hundred km its water is almost pure lake water. Seventy-seven species of fish were collected from six localities along the Lukuga River. Species of cichlids, cyprinids, and clupeids otherwise known only from Lake Tanganyika were identified from rapids in the Lukuga River at Niemba, 100 km from the lake, whereas downstream localities represent a Congo River fish fauna. Cichlid species from Niemba include special- ized algal browsers that also occur in the lake (Simochromis babaulti, S. diagramma) and one invertebrate picker representing a new species of a genus (Tanganicodus) otherwise only known from the lake. Other fish species from Niemba include an abundant species of clupeid, Stolothrissa tanganicae, otherwise only known from Lake Tangan- yika that has a pelagic mode of life in the lake. These species demonstrate that their adaptations are not neces- sarily dependent upon the lake habitat. Other endemic taxa occurring at Niemba are known to frequent vegetat- ed shore habitats or river mouths similar to the conditions at the entrance of the Lukuga, viz. Chelaethiops minutus (Cyprinidae), Lates mariae (Latidae), Mastacembelus cunningtoni (Mastacembelidae), Astatotilapia burtoni, Ctenochromis horei, Telmatochromis dhonti, and Tylochromis polylepis (Cichlidae). The Lukuga frequently did not serve as an ef- fluent due to weed masses and sand bars building up at the exit, and low water levels of Lake Tanganyika.