Sweave User Manual
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sweave User Manual
Sweave User Manual Friedrich Leisch and R-core June 30, 2017 1 Introduction Sweave provides a flexible framework for mixing text and R code for automatic document generation. A single source file contains both documentation text and R code, which are then woven into a final document containing • the documentation text together with • the R code and/or • the output of the code (text, graphs) This allows the re-generation of a report if the input data change and documents the code to reproduce the analysis in the same file that contains the report. The R code of the complete 1 analysis is embedded into a LATEX document using the noweb syntax (Ramsey, 1998) which is usually used for literate programming Knuth(1984). Hence, the full power of LATEX (for high- quality typesetting) and R (for data analysis) can be used simultaneously. See Leisch(2002) and references therein for more general thoughts on dynamic report generation and pointers to other systems. Sweave uses a modular concept using different drivers for the actual translations. Obviously different drivers are needed for different text markup languages (LATEX, HTML, . ). Several packages on CRAN provide support for other word processing systems (see Appendix A). 2 Noweb files noweb (Ramsey, 1998) is a simple literate-programming tool which allows combining program source code and the corresponding documentation into a single file. A noweb file is a simple text file which consists of a sequence of code and documentation segments, called chunks: Documentation chunks start with a line that has an at sign (@) as first character, followed by a space or newline character. -

The Pretzel User Manual
The PretzelBook second edition Felix G¨artner June 11, 1998 2 Contents 1 Introduction 5 1.1 Do Prettyprinting the Pretzel Way . 5 1.2 History . 6 1.3 Acknowledgements . 6 1.4 Changes to second Edition . 6 2 Using Pretzel 7 2.1 Getting Started . 7 2.1.1 A first Example . 7 2.1.2 Running Pretzel . 9 2.1.3 Using Pretzel Output . 9 2.2 Carrying On . 10 2.2.1 The Two Input Files . 10 2.2.2 Formatted Tokens . 10 2.2.3 Regular Expressions . 10 2.2.4 Formatted Grammar . 11 2.2.5 Prettyprinting with Format Instructions . 12 2.2.6 Formatting Instructions . 13 2.3 Writing Prettyprinting Grammars . 16 2.3.1 Modifying an existing grammar . 17 2.3.2 Writing a new Grammar from Scratch . 17 2.3.3 Context Free versus Context Sensitive . 18 2.3.4 Available Grammars . 19 2.3.5 Debugging Prettyprinting Grammars . 19 2.3.6 Experiences . 21 3 Pretzel Hacking 23 3.1 Adding C Code to the Rules . 23 3.1.1 Example for Tokens . 23 3.1.2 Example for Grammars . 24 3.1.3 Summary . 25 3.1.4 Tips and Tricks . 26 3.2 The Pretzel Interface . 26 3.2.1 The Prettyprinting Scanner . 27 3.2.2 The Prettyprinting Parser . 28 3.2.3 Example . 29 3.3 Building a Pretzel prettyprinter by Hand . 30 3.4 Obtaining a Pretzel Prettyprinting Module . 30 3.4.1 The Prettyprinting Scanner . 30 3.4.2 The Prettyprinting Parser . 31 3.5 Multiple Pretzel Modules in the same Program . -

ESS — Emacs Speaks Statistics
ESS — Emacs Speaks Statistics ESS version 5.14 The ESS Developers (A.J. Rossini, R.M. Heiberger, K. Hornik, M. Maechler, R.A. Sparapani, S.J. Eglen, S.P. Luque and H. Redestig) Current Documentation by The ESS Developers Copyright c 2002–2010 The ESS Developers Copyright c 1996–2001 A.J. Rossini Original Documentation by David M. Smith Copyright c 1992–1995 David M. Smith Permission is granted to make and distribute verbatim copies of this manual provided the copyright notice and this permission notice are preserved on all copies. Permission is granted to copy and distribute modified versions of this manual under the conditions for verbatim copying, provided that the entire resulting derived work is distributed under the terms of a permission notice identical to this one. Chapter 1: Introduction to ESS 1 1 Introduction to ESS The S family (S, Splus and R) and SAS statistical analysis packages provide sophisticated statistical and graphical routines for manipulating data. Emacs Speaks Statistics (ESS) is based on the merger of two pre-cursors, S-mode and SAS-mode, which provided support for the S family and SAS respectively. Later on, Stata-mode was also incorporated. ESS provides a common, generic, and useful interface, through emacs, to many statistical packages. It currently supports the S family, SAS, BUGS/JAGS, Stata and XLisp-Stat with the level of support roughly in that order. A bit of notation before we begin. emacs refers to both GNU Emacs by the Free Software Foundation, as well as XEmacs by the XEmacs Project. The emacs major mode ESS[language], where language can take values such as S, SAS, or XLS. -

Species' Traits and Phylogenetic
Northern Michigan University NMU Commons All NMU Master's Theses Student Works 8-2018 CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES Katie Nehiba [email protected] Follow this and additional works at: https://commons.nmu.edu/theses Part of the Ecology and Evolutionary Biology Commons Recommended Citation Nehiba, Katie, "CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES" (2018). All NMU Master's Theses. 557. https://commons.nmu.edu/theses/557 This Open Access is brought to you for free and open access by the Student Works at NMU Commons. It has been accepted for inclusion in All NMU Master's Theses by an authorized administrator of NMU Commons. For more information, please contact [email protected],[email protected]. CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES By Katie R. Nehiba THESIS Submitted to Northern Michigan University In partial fulfillment of the requirements For the degree of MASTER OF SCIENCE Office of Graduate Education and Research July 2018 ABSTRACT By Katie R. Nehiba Current anthropogenically-driven climate change is accelerating at an unprecedented rate. In response, species’ ranges may shift, tracking optimal climatic conditions. Species-specific differences may produce predictable differences in the extent of range shifts. I evaluated if patterns of predicted responses to climate change were strongly related to species’ taxonomic identities and/or ecological characteristics of species’ niches, elevation and precipitation. I evaluated differences in predicted range shifts in well-sampled small mammals that are restricted to North America: kangaroo rats, voles, chipmunks, and ground squirrels. -

Reproducible Research.. Using Sweave, Knitr and Pandoc
Reproducible Research.. Using Sweave, Knitr and Pandoc Aedín Culhane [email protected] Nov 20th 2012 My R Course Website http://bcb.dfci.harvard.edu/~aedin/ My HSPH homepage http://www.hsph.harvard.edu/research/aedin-culhane/ When issues of reproducibility arise • ``Remember that microarray analysis you did six months ago? We ran a few more arrays. Can you add them to the project and repeat the same analysis?'' • ``The statistical analyst who looked at the data I generated previously is no longer available. Can you get someone else to analyze my new data set using the same methods (and thus producing a report I can expect to understand)?'' • ``Please write/edit the methods sections for the abstract/paper/grant proposal I am submitting based on the analysis you did several months ago.'' From Keith Baggerly • Selected articles published in Nature Genetics between January 2005 and December 2006 that had used profiling with microarrays • Of the 56 items retrieved electronically, 20 articles were considered potentially eligible for the project • The four teams were from – University of Alabama at Birmingham (UAB) – Stanford/Dana-Farber (SD) – London (L) and Ioannina/Trento (IT) • Each team was comprised of 3-6 scientists who worked together to evaluate each article. Results • Result could be reproduced n=2 • Reproduced with discrepancy n=6 • Could not be reproduced n=10 – No data n=4 (no data n=2, subset n=1, no reporter data n=1) – Confusion over matching of data to analysis (n=2) – Specialized software required and not available (n=1)m – Raw data available but could not be processed n=2 Reproducibility of Analysis Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, et al, (2009) Repeatability of published microarray gene expression Analyses. -
Dynamic Documents with R and Knitr, by Yihui Xie 116 Tugboat, Volume 35 (2014), No
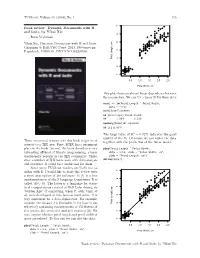
TUGboat, Volume 35 (2014), No. 1 115 ● ● ● Book review: Dynamic Documents with R ● ● ● and knitr, by Yihui Xie ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Boris Veytsman ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Yihui Xie, Dynamic Documents with R and knitr. ● ● ● ● ● ● ● ● ● ● ● ● ● ● Chapman & Hall/CRC Press, 2013, 190+xxvi pp. ● ● ● ● ● ● ● ● ● US$ ISBN ● Paperback, 59.95. 978-1482203530. ● ● ● ● Petal Length, cm Petal ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 1 2 3 4 5 6 7 0.5 1.0 1.5 2.0 2.5 Petal Width, cm This plot shows an almost linear dependence between the parameters. We can try a linear fit for these data: model <- lm(Petal.Length ˜ Petal.Width, data = iris) model$coefficients ## (Intercept) Petal.Width ## 1.084 2.230 summary(model)$r.squared ## [1] 0.9271 The large value of R2 = 0.9271 indicates the good quality of the fit. Of course we can replot the data There are several reasons why this book might be of together with the prediction of the linear model: interest to a TEX user. First, LATEX has a prominent place in the book. Second, the book describes a very plot(Petal.Length ˜ Petal.Width, interesting offshoot of literate programming, a topic data = iris, xlab = "Petal Width, cm", traditionally popular in the TEX community. Third, ylab = "Petal Length, cm") abline(model) since a number of TEX users work with data analysis and statistics, R could be a useful tool for them. Since some TUGboat readers are likely not fa- ● ● ● miliar with R, I would like to start this review with ● ● a short description of the software. R [1] is a free ● ● ● ● ● ● ● ● ● ● ● implementation of the S language (sometimes R is ● ● ● ● ● ● ● ● ● ● ● ● ● ● called GNU S). -

LYX and Knitr
knitr.1 LYX and knitr The knitr package allows one to embed R code within LYX and LATEX documents. When a document is compiled into a PDF, LYX/LATEX connects to R to run the R code and the code/output is automatically put into the PDF. In addition to this being a convenient way to use both LYX/LATEX with R, it also provides an important component to the reproducibility of research (RR). For example, one can include the code for a data analysis de- scribed in a paper. This ensures that there would be no “copying and pasting errors” and also provide readers of the paper an im- mediate way to reproduce the research. RR continues to become more important and fortunately more tools are being developed to make it possible. Below are some discussions on the topic: • AMSTAT News column on RR at http://magazine. amstat.org/blog/2011/01/01/scipolicyjan11. • CRAN task view for RR and R at http://cran.r-project. org/web/views/ReproducibleResearch.html. • Yihui Xie: Author of knitr – First and second editions of his Dynamic Documents with R and knitr book. Note that this book was typed in LYX. – Website for knitr at http://yihui.name/knitr The Sweave environment is another way to include R code inside of LYX/LATEX. This was developed prior to knitr, but it is more difficult to use. The purpose of this section is to examine the main compo- nents of knitr so that you will be able to complete the rest of the semester using LYX and knitr together for all assign- ments in our course! Also, a very important purpose is to give you the tools needed to complete all assignments in other R- based courses by using knitr and LYX together! The files used knitr.2 here are intro_example_cereal.lyx, intro_example_cereal.pdf, cereal.csv, ExternalCode.R, FirstBeamer-knitr.zip, JSM2015.zip, and RMarkdown.zip. -

Book of Abstracts
Book of Abstracts June 27, 2015 1 Conference Sponsors Diamond Sponsor Platinum Sponsors Gold Sponsors Silver Sponsors Open Analytics Bronze Sponsors Media Sponsors 2 Conference program Time Tuesday Wednesday Thursday Friday 08:00 Registration opens Registration opens Registration opens Registration opens 08:30 – 09:00 Opening session (by Rector peR! M. Johansen, Aalborg University) Aalborghallen 09:00 – 10:00 Romain François Di Cook Thomas Lumley Aalborghallen Aalborghallen Aalborghallen 10:00 – 10:30 Coffee break Coffee break Coffee break (15 min) ee break Sponsored by Quantide Sponsored by Alteryx ff Session 1 Session 4 10:30 – 12:00 Sponsor session (10:15) Kaleidoscope 1 Kaleidoscope 4 Aalborghallen Aalborghallen Aalborghallen incl. co Morning Tutorials DataRobot Ecology Medicine Gæstesalen Gæstesalen RStudio Teradata Networks Regression Musiksalen Musiksalen Revolution Analytics Reproducibility Commercial Offerings alteryx Det Lille Teater Det Lille Teater TIBCO H O Interfacing Interactive graphics 2 Radiosalen Radiosalen HP 12:00 – 13:00 Sandwiches Lunch (standing buffet) Lunch (standing buffet) Break: 12:00 – 12:30 Sponsored by Sponsored by TIBCO ff Revolution Analytics Ste en Lauritzen (12:30) Aalborghallen Session 2 Session 5 13:00 – 14:30 13:30: Closing remarks Kaleidoscope 2 Kaleidoscope 5 Aalborghallen Aalborghallen 13:45: Grab ’n go lunch 14:00: Conference ends Case study Teaching 1 Gæstesalen Gæstesalen Clustering Statistical Methodology 1 Musiksalen Musiksalen ee break Data Management Machine Learning 1 ff Det Lille Teater Det Lille -

Introduction
1 Introduction A compiler translates source code to assembler or object code for a target machine. A retargetable compiler has multiple targets. Machine-specific compiler parts are isolated in modules that are easily replaced to target different machines. This book describes lcc, a retargetable compiler for ANSI C; it fo- cuses on the implementation. Most compiler texts survey compiling al- gorithms, which leaves room for only a toy compiler. This book leaves the survey to others. It tours most of a practical compiler for full ANSI C, including code generators for three target machines. It gives only enough compiling theory to explain the methods that it uses. 1.1 Literate Programs This book not only describes the implementation of lcc,itis the imple- mentation. The noweb system for “literate programming” generates both the book and the code for lcc from a single source. This source con- sists of interleaved prose and labelled code fragments. The fragments are written in the order that best suits describing the program, namely the order you see in this book, not the order dictated by the C program- ming language. The program noweave accepts the source and produces the book’s typescript, which includes most of the code and all of the text. The program notangle extracts all of the code, in the proper order for compilation. Fragments contain source code and references to other fragments. Fragment definitions are preceded by their labels in angle brackets. For example, the code ≡ a fragment label 1 ĭ2 sum=0; for (i = 0; i < 10; i++) increment sum 1 increment sum 1≡ 1 sum += x[i]; sums the elements of x. -

Literate Programming in Java Section 1 Is Always a Starred Section
304 TUGboat, Volume 23 (2002), No. 3/4 called limbo; in this case, the introduction.) Most Software & Tools sections consist of a short text part followed by a short code part. Some sections (such as this one) contain only text, some others contain only code. Rambutan: Literate programming in Java Section 1 is always a starred section. That just means it has a title: ‘Computing primes’ in this Prasenjit Saha case. The title is supposed to describe a large group Introduction. Rambutan is a literate program- of consecutive sections, and gets printed at the start and on the page headline. Long programs have ming system for Java with TEX, closely resembling CWEB and the original WEB system.* I developed it many starred sections, which behave like chapter using Norman Ramsey’s Spidery WEB. headings. This article is also the manual, as well as an The source for this section begins example of a Rambutan literate program; that @* Computing primes. This is... is to say, the file Manual.w consists of code and In the source, @* begins a starred section, and any documentation written together in the Rambutan text up to the first period makes up the title. idiom. From this common source, the Rambutan system does two things: 2. This is an ordinary (i.e., unstarred) section, javatangle Manual and its source begins extracts a compilable Java applet to compute the @ This is an ordinary... first N primes, and In the source, @ followed by space or tab or newline javaweave Manual begins an ordinary section. In the next section things get more interesting. -

The R Journal, June 2012
The Journal Volume 2/1, June 2010 A peer-reviewed, open-access publication of the R Foundation for Statistical Computing Contents Editorial..................................................3 Contributed Research Articles IsoGene: An R Package for Analyzing Dose-response Studies in Microarray Experiments..5 MCMC for Generalized Linear Mixed Models with glmmBUGS ................. 13 Mapping and Measuring Country Shapes............................... 18 tmvtnorm: A Package for the Truncated Multivariate Normal Distribution........... 25 neuralnet: Training of Neural Networks............................... 30 glmperm: A Permutation of Regressor Residuals Test for Inference in Generalized Linear Models.................................................. 39 Online Reproducible Research: An Application to Multivariate Analysis of Bacterial DNA Fingerprint Data............................................ 44 Two-sided Exact Tests and Matching Confidence Intervals for Discrete Data.......... 53 Book Reviews A Beginner’s Guide to R......................................... 59 News and Notes Conference Review: The 2nd Chinese R Conference........................ 60 Introducing NppToR: R Interaction for Notepad++......................... 62 Changes in R 2.10.1–2.11.1........................................ 64 Changes on CRAN............................................ 72 News from the Bioconductor Project.................................. 85 R Foundation News........................................... 86 2 The Journal is a peer-reviewed publication of -

Literate Programming and Reproducible Research
JSS Journal of Statistical Software January 2012, Volume 46, Issue 3. http://www.jstatsoft.org/ A Multi-Language Computing Environment for Literate Programming and Reproducible Research Eric Schulte Dan Davison University of New Mexico Counsyl Thomas Dye Carsten Dominik University of Hawai`i University of Amsterdam Abstract We present a new computing environment for authoring mixed natural and com- puter language documents. In this environment a single hierarchically-organized plain text source file may contain a variety of elements such as code in arbitrary program- ming languages, raw data, links to external resources, project management data, working notes, and text for publication. Code fragments may be executed in situ with graphical, numerical and textual output captured or linked in the file. Export to LATEX, HTML, LATEX beamer, DocBook and other formats permits working reports, presentations and manuscripts for publication to be generated from the file. In addition, functioning pure code files can be automatically extracted from the file. This environment is implemented as an extension to the Emacs text editor and provides a rich set of features for authoring both prose and code, as well as sophisticated project management capabilities. Keywords: literate programming, reproducible research, compendium, WEB, Emacs. 1. Introduction There are a variety of settings in which it is desirable to mix prose, code, data, and compu- tational results in a single document. Scientific research increasingly involves the use of computational tools.