Near-Infrared Dual Bioluminescence Imaging in Mouse Models of Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

William D. Mcelroy Papers
http://oac.cdlib.org/findaid/ark:/13030/kt6489r0v5 No online items William D. McElroy Papers Special Collections & Archives, UC San Diego Special Collections & Archives, UC San Diego Copyright 2005 9500 Gilman Drive La Jolla 92093-0175 [email protected] URL: http://libraries.ucsd.edu/collections/sca/index.html William D. McElroy Papers MSS 0483 1 Descriptive Summary Languages: English Contributing Institution: Special Collections & Archives, UC San Diego 9500 Gilman Drive La Jolla 92093-0175 Title: William D. McElroy Papers Identifier/Call Number: MSS 0483 Physical Description: 10.4 Linear feet(19 archives boxes, 11 oversize folders, 6 art bin items) Date (inclusive): 1944-1999 Abstract: Papers of William David McElroy (1917-1999), professor of biochemistry, the fourth chancellor (1972-1980) of the University of California, San Diego; and former director (1969-1971) of the National Science Foundation. Scope and Content of Collection Papers of William David McElroy (1917-1999), professor of biochemistry, the fourth chancellor (1972-1980) of the University of California, San Diego; and former director (1969-1971) of the National Science Foundation. McElroy's significant contributions to biology include isolating and crystallizing the compounds that enable firefly luminescence and for his subsequent research into bacterial bioluminescence. He also wrote, spoke, and worked on problems in areas of environment, pollution, food production, science education, and international science. The papers largely document McElroy's scientific research and include correspondence with the scientific community, various biographical materials including awards and photographs, his trip to China in 1979, writings and reprints related to biochemical and scientific investigation, research materials on bioluminescence, teaching materials, and speeches given both as chancellor and as director of the National Science Foundation. -

Safety Data Sheet
G-Biosciences, St Louis, MO, USA | 1-800-628-7730 | 1-314-991-6034 | [email protected] A Geno Technology, Inc. (USA) brand name Safety Data Sheet Cat. # RC-227 D-Luciferin Firefly, FREE ACID Size: 0.25g think proteins! think G-Biosciences! www.GBiosciences.com D-Luciferin Firefly, potassium salt Safety Data Sheet according to Federal Register / Vol. 77, No. 58 / Monday, March 26, 2012 / Rules and Regulations Date of issue: 05/06/2013 Revision date: 05/11/2017 Version: 7.1 SECTION 1: Identification 1.1. Identification Product form : Substance Trade name : D-Luciferin Firefly, potassium salt CAS-No. : 2591-17-5 Product code : 006A Formula : C11H8N2O3S2 Synonyms : (4S)-4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-4-thiazolecarboxylic acid / (S)-2-(6-hydroxy-2- benzothiazolyl)-2-thiazoline-4-carboxylic acid / (S)-4,5-dihydro-2-(6-hydroxybenzothiazol-2- yl)thiazole-4-carboxylic acid / 2-(6-hydroxy-2-benzothiazolyl)-2-thiazoline-4-carcoxylic acid, (S)- / 2-(6-hydroxybenzothiazol-2-yl)-2-thiazoline-4-carboxylic acid / 4,5-dihydro-2-(6-hydroxy-2- benzorhiazolyl)-4-thiazolecarboxylic acid, (4S)- / 4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-4- thiazolecarboxylic acid / 4-Thiazolecarboxylic acid, 4,5-dihydro-2-(6-hydroxy-2-benzothiazolyl)-, (S)- / D-(-)-luciferin / D-luciferin / firefly luciferin / liciferin, D-(-)- / luciferin / luciferin, D- Other means of identification : D-Luciferin Firefly, free acid 4,5-Dihydro-2-(6-hydroxy-2-benzothiazolyl)-4-thiazolecarboxylic acid, ST50405784, Luciferin, CHEBI:17165 BIG No : 48631 1.2. Recommended use and restrictions on use Use of the substance/mixture : Luciferin is a common bioluminescent reporter used for in-vivo imaging of the expression of the luc marker gene . -

Review on Bioluminescence Imaging
DOI: 10.37421/apn.2020.05.170 Review Article Journal of Volume 12:7, 2021 Nuclear Medicine & Radiation Therapy ISSN: 2155-9619 Open Access Review on Bioluminescence Imaging Cleodora Yen* Department of Radiology, Yale University, United States Bioluminescence imaging (BLI) is an innovation created over the previous decade The strategy expects luciferin to be added to the circulatory system, which conveys it that takes into consideration the noninvasive investigation of continuous organic cycles. to cells all through the body. At the point when luciferin arrives at cells that have been As of late, bioluminescence tomography (BLT) has become conceivable and a few adjusted to convey the firefly quality, those cells produce light [2]. frameworks have opened up. In 2011, PerkinElmer gained perhaps the most famous lines of optical imaging frameworks with bioluminescence from Caliper Life Sciences. The BLT opposite issue of 3D remaking of the dispersion of bioluminescent atoms Bioluminescence is the cycle of light outflow in living life forms. Bioluminescence imaging from information estimated on the creature surface is innately poorly presented. The uses local light outflow from one of a few organic entities which bioluminesce. The three main little creature study utilizing BLT was directed by scientists at the University of principle sources are the North American firefly, the ocean pansy (and related marine Southern California, Los Angeles, USA in 2005. Following this turn of events, many organic entities), and microbes like Photorhabdus luminescens and Vibrio fischeri. The exploration bunches in USA and China have fabricated frameworks that empower BLT. DNA encoding the iridescent protein is fused into the lab creature either by means of Mustard plants have had the quality that makes fireflies' tails shine added to them so the a viral vector or by making a transgenic creature. -

Bioluminescence Is Produced by a Firefly-Like Luciferase but an Entirely
www.nature.com/scientificreports OPEN New Zealand glowworm (Arachnocampa luminosa) bioluminescence is produced by a Received: 8 November 2017 Accepted: 1 February 2018 frefy-like luciferase but an entirely Published: xx xx xxxx new luciferin Oliver C. Watkins1,2, Miriam L. Sharpe 1, Nigel B. Perry 2 & Kurt L. Krause 1 The New Zealand glowworm, Arachnocampa luminosa, is well-known for displays of blue-green bioluminescence, but details of its bioluminescent chemistry have been elusive. The glowworm is evolutionarily distant from other bioluminescent creatures studied in detail, including the frefy. We have isolated and characterised the molecular components of the glowworm luciferase-luciferin system using chromatography, mass spectrometry and 1H NMR spectroscopy. The purifed luciferase enzyme is in the same protein family as frefy luciferase (31% sequence identity). However, the luciferin substrate of this enzyme is produced from xanthurenic acid and tyrosine, and is entirely diferent to that of the frefy and known luciferins of other glowing creatures. A candidate luciferin structure is proposed, which needs to be confrmed by chemical synthesis and bioluminescence assays. These fndings show that luciferases can evolve independently from the same family of enzymes to produce light using structurally diferent luciferins. Glowworms are found in New Zealand and Australia, and are a major tourist attraction at sites located across both countries. In contrast to luminescent beetles such as the frefy (Coleoptera), whose bioluminescence has been well characterised (reviewed by ref.1), the molecular details of glowworm bioluminescence have remained elusive. Tese glowworms are the larvae of fungus gnats of the genus Arachnocampa, with eight species endemic to Australia and a single species found only in New Zealand2. -

Integration of Circadian and Phototransduction Pathways in the Network Controlling CAB Gene Transcription in Arabidopsis
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 15491–15496, December 1996 Plant Biology Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis (firefly luciferaseyluminescence imagingycircadian rhythmygene expression) ANDREW J. MILLAR* AND STEVE A. KAY National Science Foundation Center for Biological Timing, Department of Biology, University of Virginia, Charlottesville, VA 22903 Communicated by Winslow Briggs, Carnegie Institute of Washington, Stanford, CA, October 15, 1996 (received for review June 24, 1996) ABSTRACT The transcription of CAB genes, encoding the the circadian clock acts negatively on CAB expression, antag- chlorophyll ayb-binding proteins, is rapidly induced in dark- onizing the positive effects of phototransduction pathways grown Arabidopsis seedlings following a light pulse. The during the subjective night. transient induction is followed by several cycles of a circadian The transient induction of CAB expression immediately rhythm. Seedlings transferred to continuous light are known after plants are exposed to light is referred to as the ‘‘acute’’ to exhibit a robust circadian rhythm of CAB expression. The response to light (9, 11). Many circadian-regulated processes precise waveform of CAB expression in light–dark cycles, exhibit acute effects of light: the expression of clock-controlled however, reflects a regulatory network that integrates infor- genes (ccg)inNeurospora is induced by light (12), for example, mation from photoreceptors, from the circadian clock and and the production of melatonin in cultured avian pinealocytes possibly from a developmental program. We have used the is suppressed (13). Current evidence indicates that the circa- luciferase reporter system to investigate CAB expression with dian oscillator is not required for these acute responses: ccg high time resolution. -
The Colors of Firefly Bioluminescence: Enzyme Configuration and Species Specificity by H
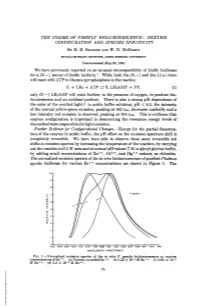
THE COLORS OF FIREFLY BIOLUMINESCENCE: ENZYME CONFIGURATION AND SPECIES SPECIFICITY BY H. H. SELIGER AND W. D. MCELROY MCCOLLUM-PRATT INSTITUTE, JOHNS HOPKINS UNIVERSITY Communicated May 25, 1964 We have previously reported on an unusual stereospecificity of firefly luciferase for a D(-) isomer of firefly luciferin.' While both the D(-) and the L(+) form will react with ATP to liberate pyrophosphate in the reaction E + LH2 + ATP =- E. LH2AMP + PP, (1) only D(-) LH2AMP will react further, in the presence of oxygen, to produce bio- luminescence and an oxidized product. There is also a strong pH dependence of the color of the emitted light;2 in acidic buffer solutions, pH < 6.5, the intensity of the normal yellow-green emission, peaking at 562 ml,, decreases markedly and a low intensity red emission is observed, peaking at 616 miu. This is evidence that enzyme configuration is important in determining the resonance energy levels of the excited state responsible for light emission. Further Evidence for Configurational Changes.-Except for the partial denatura- tion of the enzyme in acidic buffer, the pH effect on the emission spectrum shift is completely reversible. We have been able to observe these same reversible red shifts in emission spectra by increasing the temperature of the reaction, by carrying out the reaction in 0.2 M urea and at normal pH values (7.6) in glycyl glycine buffer, by adding small concentrations of Zn++, Cd++, and Hg++ cations, as chlorides. The normalized emission spectra of the in vitro bioluminescence of purified Photinus pyralis luciferase for various Zn++ concentrations are shown in Figure 1. -

Bioluminescence in Insect
Int.J.Curr.Microbiol.App.Sci (2018) 7(3): 187-193 International Journal of Current Microbiology and Applied Sciences ISSN: 2319-7706 Volume 7 Number 03 (2018) Journal homepage: http://www.ijcmas.com Review Article https://doi.org/10.20546/ijcmas.2018.703.022 Bioluminescence in Insect I. Yimjenjang Longkumer and Ram Kumar* Department of Entomology, Dr. Rajendra Prasad Central Agricultural University, Pusa, Bihar-848125, India *Corresponding author ABSTRACT Bioluminescence is defined as the emission of light from a living organism K e yw or ds that performs some biological function. Bioluminescence is one of the Fireflies, oldest fields of scientific study almost dating from the first written records Bioluminescence , of the ancient Greeks. This article describes the investigations of insect Luciferin luminescence and the crucial role imparted in the activities of insect. Many Article Info facets of this field are easily accessible for investigation without need for Accepted: advanced technology and so, within the History of Science, investigations 04 February 2018 of bioluminescence played a significant role in the establishment of the Available Online: scientific method, and also were among the many visual phenomena to be 10 March 2018 accounted for in developing a theory of light. Introduction Bioluminescence (BL) serves various purposes, including sexual attraction and When a living organism produces and emits courtship, predation and defense (Hastings and light as a result of a chemical reaction, the Wilson, 1976). This process is suggested to process is known as Bioluminescence - bio have arisen after O2 appearance on Earth at means 'living' in Greek while `lumen means least 30 different times during evolution, as 'light' in Latin. -

Determining the Depth Limit of Bioluminescent Sources in Scattering Media
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.21.044982; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Determining the Depth Limit of Bioluminescent Sources in Scattering Media. Ankit Raghuram1,*, Fan Ye1, Jesse K. Adams1,2, Nathan Shaner3, Jacob T. Robinson1,2,4,5, Ashok Veeraraghavan1,2,6 1 Department of Electrical and Computer Engineering, Rice University, Houston, TX 77005, USA 2 Applied Physics Program, Rice University, Houston, TX 77005, USA 3 Department of Neurosciences, University of California San Diego, La Jolla, CA 92093, USA 4 Department of Neuroscience, Baylor College of Medicine, Houston, TX 77030, USA 5 Department of Bioengineering, Rice University, Houston, TX 77005, USA 6 Department of Computer Science, Rice University, Houston TX 77005, USA * [email protected] Abstract Bioluminescence has several potential advantages compared to fluorescence microscopy for in vivo biological imaging. Because bioluminescence does not require excitation light, imaging can be performed for extended periods of time without phototoxicity or photobleaching, and optical systems can be smaller, simpler, and lighter. Eliminating the need for excitation light may also affect how deeply one can image in scattering biological tissue, but the imaging depth limits for bioluminescence have yet to be reported. Here, we perform a theoretical study of the depth limits of bioluminescence microscopy and find that cellular resolution imaging should be possible at a depth of 5-10 mean free paths (MFPs). This limit is deeper than the depth limit for confocal microscopy and slightly lower than the imaging limit expected for two-photon microscopy under similar conditions. -

Bioluminescence Imaging of DNA Synthetic Phase of Cell Cycle in Living Animals
Bioluminescence Imaging of DNA Synthetic Phase of Cell Cycle in Living Animals Zhi-Hong Chen1,2, Rui-Jun Zhao1,3, Rong-Hui Li1,3, Cui-Ping Guo1,3, Guo-Jun Zhang1,3* 1 Breast Center, Cancer Hospital of Shantou University Medical College, Shantou, People’s Republic of China, 2 Heilongjiang Province Key Laboratory of Cancer Prevention and Treatment, Mudanjiang Medical University, Mudanjiang, People’s Republic of China, 3 Cancer Research Center, Shantou University Medical College, Shantou, People’s Republic of China Abstract Bioluminescence reporter proteins have been widely used in the development of tools for monitoring biological events in living cells. Currently, some assays like flow cytometry analysis are available for studying DNA synthetic phase (S-phase) targeted anti-cancer drug activity in vitro; however, techniques for imaging of in vivo models remain limited. Cyclin A2 is known to promote S-phase entry in mammals. Its expression levels are low during G1-phase, but they increase at the onset of S-phase. Cyclin A2 is degraded during prometaphase by ubiquitin-dependent, proteasome-mediated proteolysis. In this study, we have developed a cyclin A2-luciferase (CYCA-Luc) fusion protein targeted for ubiquitin-proteasome dependent degradation, and have evaluated its utility in screening S-phase targeted anti-cancer drugs. Similar to endogenous cyclin A2, CYCA-Luc accumulates during S-phase and is degraded during G2/M-phase. Using Cdc20 siRNA we have demonstrated that Cdc20 can mediate CYCA-Luc degradation. Moreover, using noninvasive bioluminescent imaging, we demonstrated accumulation of CYCA-Luc in response to 10-hydroxycamptothecin (HCPT), an S-phase targeted anti-cancer drug, in human tumor cells in vivo and in vitro. -

Brazilian Bioluminescent Beetles: Reflections on Catching Glimpses of Light in the Atlantic Forest and Cerrado
Anais da Academia Brasileira de Ciências (2018) 90(1 Suppl. 1): 663-679 (Annals of the Brazilian Academy of Sciences) Printed version ISSN 0001-3765 / Online version ISSN 1678-2690 http://dx.doi.org/10.1590/0001-3765201820170504 www.scielo.br/aabc | www.fb.com/aabcjournal Brazilian Bioluminescent Beetles: Reflections on Catching Glimpses of Light in the Atlantic Forest and Cerrado ETELVINO J.H. BECHARA and CASSIUS V. STEVANI Departamento de Química Fundamental, Instituto de Química, Universidade de São Paulo, Av. Prof. Lineu Prestes, 748, 05508-000 São Paulo, SP, Brazil Manuscript received on July 4, 2017; accepted for publication on August 11, 2017 ABSTRACT Bioluminescence - visible and cold light emission by living organisms - is a worldwide phenomenon, reported in terrestrial and marine environments since ancient times. Light emission from microorganisms, fungi, plants and animals may have arisen as an evolutionary response against oxygen toxicity and was appropriated for sexual attraction, predation, aposematism, and camouflage. Light emission results from the oxidation of a substrate, luciferin, by molecular oxygen, catalyzed by a luciferase, producing oxyluciferin in the excited singlet state, which decays to the ground state by fluorescence emission. Brazilian Atlantic forests and Cerrados are rich in luminescent beetles, which produce the same luciferin but slightly mutated luciferases, which result in distinct color emissions from green to red depending on the species. This review focuses on chemical and biological aspects of Brazilian luminescent beetles (Coleoptera) belonging to the Lampyridae (fireflies), Elateridae (click-beetles), and Phengodidae (railroad-worms) families. The ATP- dependent mechanism of bioluminescence, the role of luciferase tuning the color of light emission, the “luminous termite mounds” in Central Brazil, the cooperative roles of luciferase and superoxide dismutase against oxygen toxicity, and the hypothesis on the evolutionary origin of luciferases are highlighted. -

Strongly Enhanced Bacterial Bioluminescence with the Ilux Operon for Single-Cell Imaging
Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging Carola Gregora,1, Klaus C. Gwoscha, Steffen J. Sahla, and Stefan W. Hella,b,1 aDepartment of NanoBiophotonics, Max Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany; and bDepartment of Optical Nanoscopy, Max Planck Institute for Medical Research, 69120 Heidelberg, Germany Contributed by Stefan W. Hell, December 8, 2017 (sent for review September 12, 2017; reviewed by David W. Piston and Alice Y. Ting) Bioluminescence imaging of single cells is often complicated by the filters are required, making the setup very simple. In addition, it is requirement of exogenous luciferins that can be poorly cell- possible to study processes where the intense excitation light re- permeable or produce high background signal. Bacterial biolumi- quired for fluorescence measurements would be disturbing, such + nescence is unique in that it uses reduced flavin mononucleotide as circadian rhythms or Ca2 activity in the retina (3, 4). Third, no as a luciferin, which is abundant in all cells, making this system phototoxicity or bleaching occurs, allowing image acquisition over purely genetically encodable by the lux operon. Unfortunately, the arbitrary timespans. Furthermore, bioluminescence is dependent use of bacterial bioluminescence has been limited by its low on metabolic energy, and hence, only metabolically active cells are brightness compared with other luciferases. Here, we report the visible, preventing artifacts due to the observation of severely generation of an improved lux operon named ilux with an approx- damaged or dead cells. imately sevenfold increased brightness when expressed in Escheri- In addition to the limitation by their low brightness, the lucif- chia coli; ilux can be used to image single E. -

(12) United States Patent (10) Patent No.: US 9,574.223 B2 Cali Et Al
USO095.74223B2 (12) United States Patent (10) Patent No.: US 9,574.223 B2 Cali et al. (45) Date of Patent: *Feb. 21, 2017 (54) LUMINESCENCE-BASED METHODS AND 4,826,989 A 5/1989 Batz et al. PROBES FOR MEASURING CYTOCHROME 4,853,371 A 8/1989 Coy et al. 4,992,531 A 2f1991 Patroni et al. P450 ACTIVITY 5,035,999 A 7/1991 Geiger et al. 5,098,828 A 3/1992 Geiger et al. (71) Applicant: PROMEGA CORPORATION, 5,114,704 A 5/1992 Spanier et al. Madison, WI (US) 5,283,179 A 2, 1994 Wood 5,283,180 A 2f1994 Zomer et al. 5,290,684 A 3/1994 Kelly (72) Inventors: James J. Cali, Verona, WI (US); Dieter 5,374,534 A 12/1994 Zomer et al. Klaubert, Arroyo Grande, CA (US); 5,498.523 A 3, 1996 Tabor et al. William Daily, Santa Maria, CA (US); 5,641,641 A 6, 1997 Wood Samuel Kin Sang Ho, New Bedford, 5,650,135 A 7/1997 Contag et al. MA (US); Susan Frackman, Madison, 5,650,289 A T/1997 Wood 5,726,041 A 3/1998 Chrespi et al. WI (US); Erika Hawkins, Madison, WI 5,744,320 A 4/1998 Sherf et al. (US); Keith V. Wood, Mt. Horeb, WI 5,756.303 A 5/1998 Sato et al. (US) 5,780.287 A 7/1998 Kraus et al. 5,814,471 A 9, 1998 Wood (73) Assignee: PROMEGA CORPORATION, 5,876,946 A 3, 1999 Burbaum et al. 5,976,825 A 11/1999 Hochman Madison, WI (US) 6,143,492 A 11/2000 Makings et al.