Ammoperdix Griseogularis, Galliformes) Populations from Sub-Himalayan Mountain Ranges of Pakistan
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Native Partridges of Turkey
The native partridges of Turkey ALPER YILMAZ * and CAFER TEPELI Department of Animal Science, Faculty of Veterinary Medicine, University of Selcuk, 42031, Selçuklu, Konya, Turkey. *Correspondence author - [email protected] Paper presented at the 4 th International Galliformes Symposium, 2007, Chengdu, China. Abstract Turkey provides a wide range of natural habitat for numerous bird species. Partridges constitute an important part of the native birds of Turkey. There are five native partridge species in Turkey, which are chukar partridge, rock partridge, grey partridge, see-see partridge and Caspian snowcock. In recent years, intensive rearing and releasing of gamebirds has become popular in Turkey and rock partridges are an important component to this activity. Breeding units for the species are widespread in many parts of the country. There are also some breeding units for rock partridges that are supported by the National Ministry of the Forest. The units produce and release partridges to bolster the wild population, but also to provide birds for hunting and tourism. In this paper the geographical distribution, characteristics and contemporary state of the native partridges of Turkey is presented. Keywords Distribution, partridges, status, Turkey. Introduction Urfa (south east of Turkey) is thought to be over 2500 years old (FIG . 1) (T.C. Şanlı Urfa Turkey occupies a unique geographical location, Valiliği İl Kültür ve Turizm Müdürlüğü, 2007). connecting Europe and Asia, and is a country with a rich and varied historical past. The country’s rich history, geography and nature are entwined and are a part of everyday life. The total number of bird species within Turkey is equal to the number within the whole of Europe due in part to the Anatolian region’s diversity of habitats, including lakes, swamps, mountains, woodlands, and its location on major bird immigration routes (Anonymous, 1986; Boyla, 1995; Somçağ, 2005). -
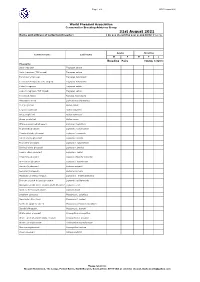
31St August 2021 Name and Address of Collection/Breeder: Do You Closed Ring Your Young Birds? Yes / No
Page 1 of 3 WPA Census 2021 World Pheasant Association Conservation Breeding Advisory Group 31st August 2021 Name and address of collection/breeder: Do you closed ring your young birds? Yes / No Adults Juveniles Common name Latin name M F M F ? Breeding Pairs YOUNG 12 MTH+ Pheasants Satyr tragopan Tragopan satyra Satyr tragopan (TRS ringed) Tragopan satyra Temminck's tragopan Tragopan temminckii Temminck's tragopan (TRT ringed) Tragopan temminckii Cabot's tragopan Tragopan caboti Cabot's tragopan (TRT ringed) Tragopan caboti Koklass pheasant Pucrasia macrolopha Himalayan monal Lophophorus impeyanus Red junglefowl Gallus gallus Ceylon junglefowl Gallus lafayettei Grey junglefowl Gallus sonneratii Green junglefowl Gallus varius White-crested kalij pheasant Lophura l. hamiltoni Nepal Kalij pheasant Lophura l. leucomelana Crawfurd's kalij pheasant Lophura l. crawfurdi Lineated kalij pheasant Lophura l. lineata True silver pheasant Lophura n. nycthemera Berlioz’s silver pheasant Lophura n. berliozi Lewis’s silver pheasant Lophura n. lewisi Edwards's pheasant Lophura edwardsi edwardsi Vietnamese pheasant Lophura e. hatinhensis Swinhoe's pheasant Lophura swinhoii Salvadori's pheasant Lophura inornata Malaysian crestless fireback Lophura e. erythrophthalma Bornean crested fireback pheasant Lophura i. ignita/nobilis Malaysian crestless fireback/Vieillot's Pheasant Lophura i. rufa Siamese fireback pheasant Lophura diardi Southern Cavcasus Phasianus C. colchicus Manchurian Ring Neck Phasianus C. pallasi Northern Japanese Green Phasianus versicolor -

Patterns of Success in Game Bird Introductions in the United States
February 18, 2016 1 Patterns of Success in Game Bird Introductions in the United States 2 3 4 by 5 6 7 8 1Michael P. Moulton, 2Wendell P. Cropper, Jr., 1Andrew J. Broz, 2Salvador A. 9 Gezan 10 11 1Department of Wildlife Ecology and Conservation; University of Florida; PO 12 Box 110430; Gainesville, FL 32611-0430; <[email protected]> 13 2School of Forest Resources and Conservation; University of Florida; PO Box 14 110410; Gainesville, FL 32611-0410; <[email protected]> 15 16 "There is no safety in numbers, or in anything else." James Thurber, The Fairly 17 Intelligent Fly, The New Yorker, February 4, 1939. 18 19 20 1 PeerJ PrePrints | https://doi.org/10.7287/peerj.preprints.1765v1 | CC-BY 4.0 Open Access | rec: 20 Feb 2016, publ: 20 Feb 2016 February 18, 2016 21 Abstract 22 Better predictions of the success of species’ introductions require careful 23 evaluation of the relative importance of at least three kinds of factors: species 24 characteristics, characteristics of the site of introduction, and event-level factors 25 such as propagule pressure. Historical records of bird introductions provide a 26 unique method for addressing the relative importance of these factors. We compiled 27 a list of introductions of 17 Phasianid species released in the USA during the Foreign 28 Game Investigation Program (FGIP). These records indicate that releases for some 29 Phasianid species in the USA continued long after establishment. For many of the 13 30 species that always failed, even numerous releases and large numbers of individuals 31 per release were not enough for successful establishment, yet several of these 32 species were successfully introduced elsewhere. -

Remembering Dr. Arthur Crane Risser by Josef Lindholm III, It Stayed That Way Through the Rest of the 1960’S
Remembering Dr. Arthur Crane Risser by Josef Lindholm III, it stayed that way through the rest of the 1960’s. On Dec. 31, Senior Aviculturist, The Dallas World Aquarium 1969, it reached an all-time high of 1,126 species and subspecies of birds (and 3,465 specimens). Then it dropped. At the end of Art Risser’s death following a stroke on the day after Christ- 1970 there were 1,097 taxa. On Jan. 1, 1972, there were 917. A mas 2008, was entirely unexpected. But many of his saddened year later there were 856. And on Jan1., 1974, the number stood friends were also startled to learn he was 70. I think most of us at 772. I found this deeply disturbing. thought he was far younger. When I first met him, shortly after At the same time, my own small avicultural world had also his arrival at the San Diego Zoo, as Assistant Curator of Birds, become much smaller. In 1972, I was, with much effort, able to in 1974, I thought he was in his late twenties. He was, in fact, 35 convince my parents to buy me Red-eared Waxbills at Wool- when he thus entered the zoo profession, having previously been worth’s and Strawberry Finches and Cut-throats at the White involved in mammalogy. Front, all for $3.95 a pair. In 1974, I found the prices for all of He earned his Master’s in Wildlife Management from the these were now $40 a pair. University of Arizona, in 1963, conducting field research on In answer to the question that all young zoo enthusiasts ask: White-nosed Coatis. -

Ammoperdix Griseogularis
Ammoperdix griseogularis -- (Brandt, 1843) ANIMALIA -- CHORDATA -- AVES -- GALLIFORMES -- PHASIANIDAE Common names: See-see Partridge; See-see European Red List Assessment European Red List Status LC -- Least Concern, (IUCN version 3.1) Assessment Information Year published: 2015 Date assessed: 2015-03-31 Assessor(s): BirdLife International Reviewer(s): Symes, A. Compiler(s): Ashpole, J., Burfield, I., Ieronymidou, C., Pople, R., Wheatley, H. & Wright, L. Assessment Rationale European regional assessment: Least Concern (LC) EU27 regional assessment: Not Applicable (NA) Although this species may have a restricted range, it is not believed to approach the thresholds for Vulnerable under the range size criterion (Extent of Occurrence 10% in ten years or three generations, or with a specified population structure). The population trend appears to be stable, and hence the species does not approach the thresholds for Vulnerable under the population trend criterion (30% decline over ten years or three generations). For these reasons the species is evaluated as Least Concern in Europe. The bird is not known from the EU27 region. Occurrence Countries/Territories of Occurrence Native: Azerbaijan; Turkey Population The European population is estimated at 4,000-6,100 pairs, which equates to 8,000-12,100 mature individuals. The species does not occur in the EU27. For details of national estimates, see Supplementary PDF . Trend In Europe the population size is estimated to be stable. For details of national estimates, see Supplementary PDF. Habitats and Ecology The species is found on dry and stony foothills that are lightly vegetated or bare, occasionally on plateaux up to approximately 2,000 m. It is found in warmer, lower areas than Alectoris chukar where they are sympatric (McGowan and Kirwan 2014). -

Management of Galliformes
38_Galliforms.qxd 8/24/2005 10:02 AM Page 861 CHAPTER 38 Management of Galliformes GARY D. BUTCHER, BS, MS, DVM, P hD, D ipl ACPV Members of the order Galliformes are found on every continent except Antarctica. The red junglefowl, com- mon turkey and helmeted guinea fowl have been domesticated for centuries. Their descendants, through selective breeding, are of considerable economic impor- tance today. Some varieties are very plentiful in the wild, while others like the Japanese quail (Coturnix japonica) and various pheasants are approaching a level of com- plete domestication. Many Galliformes are commonly maintained as game and food (meat and/or eggs) birds. Some are stable in captivity under variable ambient conditions, easy to breed and inexpensive. Other species are from niches with specific environmental requirements and need spe- cialized diets, humidity and temperature ranges to sur- vive. Currently, commercial production of chickens and turkeys in the USA for food has surpassed that of the Greg J. Harrison Greg J. Fig 38.1 | In warm climates, game cocks are housed on “string beef, pork and fish industries. In 1900, per capita con- walks”. A leg leash, just long enough for the bird to reach a sumption of chicken was 1 pound and had risen to 80 shelter but not to fight with other birds, allows group confine- pounds by the year 2000! ment. Fighting of cocks is considered inhumane and illegal in some countries; however, it is a part of the culture in others. In this chapter, “domestic fowl” means Gallus gallus, forma domestica (domestic form of the red junglefowl); “domestic turkey” is Meleagris gallopavo, forma domes- tica (domestic form of the common turkey) and “domes- tic guinea fowl” is Numida meleagris, forma domestica (domestic form of the helmeted guinea fowl) (Table 38.1). -

Parasitosis of the Chukar Partridge, Alectoris Chukarin North Iraq
240 Original Investigation / Özgün Araştırma Parasitosis of the Chukar Partridge, Alectoris chukar in North Iraq Kuzey Irak’ın Kınalı Keklik (Alectoris chukar) Parazitleri Shlemon Al-Barwari1, Isam Saeed2 1Formerly of Salahaddin University, Biology Division, Erbil, Iraq 2University Collage of Sjælland, Bioanalytiker, Næstved, Denmark ABSTRACT Objective: The aim of this study was to establish the identity of the parasitic communities of the chukar partridge from Iraq as well as report- ing on the prevalence and intensity of various infections from several localities and comprising both sexes. Methods: Twenty-nine live adult chukar partridge (Alectoris chukar) were obtained from several localities across the northern provinces of Iraq. Blood samples were examined for haemoprotozoa, carcasses were assessed for the ectoparasites throughout the body skins and feathers, and the alimentary canal was examined for protozoans and helminths. Results: All of the examined animals were infected with the 18 different endo- and ectoparasite species identified. These include 2 proto- zoans, 3 cestodes, 2 nematodes, and 11 arthropods. The overall figures suggest no significant differences in infection indices in terms of the host’s sexuality. Pathogenicity of the parasites involved is briefly emphasised. Furthermore, the role of the chukar in the dissemination of these disease agents among populations of other avian species is discussed. Conclusion: With the exception of C. latiproglottina, all of the species differentiated represent new records for chukar from Iraq, and the 2 coccidians are new addenda to the country parasitological list. (Turkiye Parazitol Derg 2012; 36: 240-6) Key Words: Chukar partridge, parasite, endoparasite, ectoparasite, Irak Received: 10.04.2012 Accepted: 02.08.2012 ÖZET Amaç: Irak’tan toplanan kınalı kekliklerde bulunan parazitlerin yaygınlığı ve yoğunluğunu belirlemek amacıyla yapılmıştır. -

A Nearly Complete Skeleton of the Fossil Galliform Bird Palaeortyx from the Late Oligocene of Germany
ACTA ORNITHOLOGICA Vol. 41 (2006) No. 2 A nearly complete skeleton of the fossil galliform bird Palaeortyx from the late Oligocene of Germany Gerald MAYR1, Markus POSCHMANN2 & Michael WUTTKE2 1Forschungsinstitut Senckenberg, Sektion für Ornithologie, Senckenberganlage 25, D-60325 Frankfurt am Main, GERMANY, e-mail: [email protected] 2Landesamt für Denkmalpflege Rheinland-Pfalz, Referat Erdgeschichtliche Denkmalpflege, Große Langgasse 29, D-55116 Mainz, GERMANY, e-mail: [email protected] Mayr G., Poschmann M., Wuttke M. 2006. A nearly complete skeleton of the fossil galliform bird Palaeortyx from the late Oligocene of Germany. Acta Ornithol. 41: 129–135. Abstract. Phasianid galliform birds do not occur in Europe before the end of the early Oligocene, and their Paleogene fossil record mainly comprises isolated bones. Here we describe a nearly complete and articulated skeleton of a phasianid galliform bird from the late Oligocene of Enspel in Germany. The specimen is assigned to Palaeortyx cf. gallica Milne-Edwards, 1869 and for the first time allows the recognition of cranial and pelvic details in a late Oligocene phasianid. Many gizzard stones are preserved in both the gizzard and the crop, and constitute the earliest fossil evidence for grit use in a phasianid galliform. Key words: fossil birds, Galliformes, Palaeortyx, gizzard stones, Oligocene Received — July 2006, accepted — Oct. 2006 INTRODUCTION pectoral girdle bones only. The only other phasianid from the Oligocene of Europe is As traditionally recognized -

Aves, Galliformes): a Cladistic Analysis of Morphological Characters
Blackwell Science, LtdOxford, UKZOJZoological Journal of the Linnean Society0024-4082The Lin- nean Society of London, 2003 137 Original Article G. J. DYKE ET AL .SUPRAGENERIC RELATIONSHIPS OF GALLIFORM BIRDS Zoological Journal of the Linnean Society, 2003, 137, 227–244. With 6 figures Suprageneric relationships of galliform birds (Aves, Galliformes): a cladistic analysis of morphological characters GARETH J. DYKE1,*, BONNIE E. GULAS2 and TIMOTHY M. CROWE1,3 1Division of Vertebrate Zoology (Ornithology), American Museum of Natural History, Central Park West at 79th Street, New York, NY 10024-5192, USA 2Department of Geology and Geophysics, Yale University, PO Box 208109, New Haven, CT 06520, USA 3Percy FitzPatrick Institute, University of Cape Town, Private Bag, Rondebosch 7701, Western Cape, South Africa Received January 2002; accepted for publication June 2002 Of the basal clades of extant birds (Neornithes) the ‘landfowl’ or galliforms (Aves, Galliformes) are the most speciose. Cladistic analysis of more than 100 morphological characters coded at the generic level for most putative galliform genera confirms that the megapodes (‘mound builders’; Megapodiidae) are the most basal clade within the order. They are followed successively by the curassows, guans and chachalacas (Cracidae), which comprise the sister-group to all other extant Galliformes (i.e. Phasianoidea). Within this large ‘phasianoid’ clade, analyses suggest that the guineafowl (Numididae) are the most basal taxon, although monophyly of this ‘family’ is not strictly supported on the basis of the morphological characters employed. An additional major clade within the phasianoid Galliformes is recovered by this analysis, comprising the traditional groupings of New World quails (Odontophoridae) and Old World quails (‘Perdicini’), yet only monophyly of the former is supported unambiguously by morphological characters. -

Distribution and Status of Galliformes in the United Arab Emirates
Distribution and status of Galliformes in the United Arab Emirates * SHAHID BASHIR KHAN , SALIM JAVED and JUNID NAZEER SHAH Terrestrial Environment Research Centre, Environment Agency – Abu Dhabi, PO Box 45553, Abu Dhabi, United Arab Emirates *Correspondence author - [email protected] Paper presented at the 4 th International Galliformes Symposium, 2007, Chengdu, China. Abstract This paper reviews the distribution and status of Galliformes in the United Arab Emirates (UAE) over recent years. Hunting being strictly forbidden, the protection offered to bird species in general and Galliformes in particular in the UAE has helped them establish healthy breeding populations and some of them have expanded their range. Five species of Galliformes occur in the UAE, these being chukar partridge, sand partridge, black francolin, grey francolin and European quail. Sand partridge is a native resident breeder and is a regional priority species due to its restricted range. Grey francolin is a resident breeder and is expanding its range. Quail is a passage migrant and has now been confirmed as resident breeder in some areas. Its presence has been reported from areas that have witnessed an increase in fodder farms. The other two species, black francolin and chukar partridge were deliberately introduced and they have now established self sustaining breeding populations in some areas, however, they have not been able to spread to new areas as efficiently as the grey francolin. Keywords Desert ecosystem, Galliformes introductions, habitat alteration, plantation, range expansion Introduction Galliformes in the country are represented by United Arab Emirates (UAE) is a confederation five species; sand partridge Ammoperdix heyi, of seven individual emirates and the country is grey francolin Francolinus pondicerianus , quail bordered by Saudi Arabia, Oman and the Coturnix coturnix , black francolin Francolinus Arabian Gulf. -

Summer Water Turnover Rates in Free-Living Chukars and Sand Partridges in the Negev Desert
Condor 85:333-337 cc>The Cooper Ornithological Society 1983 SUMMER WATER TURNOVER RATES IN FREE-LIVING CHUKARS AND SAND PARTRIDGES IN THE NEGEV DESERT A. ALLAN DEGEN BERRY PINSHOW AND PHILIP U. ALKON ABSTRACT.-The Chukar (Alectoris chukar) and the Sand Partridge (AW~MZO- perdix heyi) are permanent residents of the Negev desert, Israel. They are sym- patric between the 200 and 90 mm isohyets, with only Sand Partridges occurring in more arid areas. During the dry season,we measured total body water volume (TBW; as tritiated water space) and total water turnover rate, and estimated dry matter intake and components of water turnover in free-living birds of both species.Mean body mass(m,) of 40 Chukars was 460.8 g and of 42 Sand Partridges was 176.6 g. TBW, as a percentage of m,,, was lower in Chukars than in Sand Partridges (67.4% vs. 69.8%; P < 0.05). Mean water turnover was 44.1 ml/day for Chukars (n = 29) and 20.8 ml/day for Sand Partridges (n = 39). The mass- specificwater turnover rate was higher in Sand Partridgesthan in Chukars (122.5 ml kg-’ day-’ vs. 100.6 ml kg-’ day-‘; P < 0.05). However, when compared allometrically, per kg07s, water turnover rates were similar (8 1.7 ml kgmo.75day-l for Chukars vs. 78.5 ml kg-O.‘* day-’ for Sand Partridges). Assuming a diet of seeds,we calculated that drinking water amounted to about 70% of water turnover in both Chukars and Sand Partridges and we concluded that both speciesmust drink water when only dry forage is available. -

Genetic Diversity of Pheasants from Natural Habitat and Farm Breeding in Eastern Poland
Vol. 12(18), pp. 2313-2321, 1 May, 2013 DOI: 10.5897/AJB2012.2980 African Journal of Biotechnology ISSN 1684-5315 ©2013 Academic Journals http://www.academicjournals.org/AJB Full Length Research Paper Genetic diversity of pheasants from natural habitat and farm breeding in Eastern Poland 1 2 3 Magdalena Gryzińska *, Roman Dziedzic and Josef Feuereisel 1Department of Biological Basis of Animal Production, Faculty of Animals’ Biology and Breeding, University of Life Sciences in Lublin, Poland. 2Department of Ecology and Wildlife Management, Faculty of Animals’ Biology and Breeding, University of Life Sciences in Lublin, Poland. 3Forestry and Game Management Research Institute, Jíloviště, The Czech Republic. Accepted 4 April, 2013 The present model of wildlife management in relationship to pheasants is based on closed breeding and insertion of grown birds into the hunting grounds. The survival rate of pheasants from farm breeding does not exceed 10%. One can pose a hypothesis about the genetic determination of survivability of pheasants originating from farm breeding. A study of the DNA polymorphism of two groups of pheasants was performed. The first group comprised birds living in a natural habitat in Central-Eastern Poland, where no farm-bred pheasants had been inserted for seven years and where wildlife management was focused on the formation of good habitat conditions, an optimum nutritional base, and low-level shooting of cocks. The second group was of birds bred on a farm and from this farm, pheasants were inserted within the area of Central-Eastern Poland. Their DNA was isolated from their feathers. Amplification of fragments of DNA was with the random amplified polymorphic DNA polymrease chain reactrion (RAPD-PCR) method.