Regulation of Ribonucleotide Reductase and Screening for Species-Specific Inhibitors in Fission Yeast
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phylogenetic Screening for Possible Novel
11 M060072591U NORTH-WEST UNIVERSITY tilt• YUNIBESITI YA BOKONE•BOPHIRIMA NOOROVVE S-UNIVERSITEIT PHYLOGENETIC SCREENING FOR POSSIBLE NOVEL ANTIBIOTIC PRODUCING ACTINOMYCETES FROM RHIZOSPHERIC SOIL SAMPLES COLLECTED FROM NGAKA MODIRI MOLEMA DISTRICT IN NORTH WEST PROVINCE, SOUTH AFRICA I BY MOBOLAJI FELICIA ADEGBOYE A thesis submitted in fulfilment of the requirements for the degree of DOCTOR OF PHILOSOPHY (BIOLOGY) DEPARTMENT OF BIOLOGICAL SCIENCES FACULTY OF SCIENCE, AGRICULTURE AND TECHNOLOGY NORTH-WEST UNIVERSITY, MAFIKENG CAMPUS SOUTH AFRICA Supervisor: Professor Olubukola 0. Babalola 2014 LIBRARY o MAFIKENG CAMPUS CALL NO.: 2021 -02- 0 4 DECLARATION I, the undersigned, declare that this thesis submitted to the North-West University for the degree of Doctor of Philosophy in Biology in the Faculty of Science, Agriculture and Technology, School of Environmental and Health Sciences, and the work contained herein is my original work with exemption to the citations and that this work has not been submitted at any other University in partial or entirely for the award of any degree. Name: Mobolaji Felicia Adegboye Signature: .....~ •·· ··· ····· ·· .. ··············· ..... Date: .... ~S.. .. ....a~ ·1·· ·'.}Q~i; ... ............ .... DEDICATION This work is dedicated to Almighty God for His faithfulness over my life and for making my helpers to be many. ii ACKNOWLEDGEMENTS I would like to express my deepest thanks, gratitude and appreciation to my supervisor and mentor, Prof. Olubukola 0. Babalola for giving me the opportunity to pursue my doctoral degree under her supervision and for her encouragement, help and kind support. Her invaluable advice, suggestions, discussions and guidance were a real support to me. I acknowledge with honour and gratitude the International Foundation for Science (IFS) for research grant (F/5330-1 ), Connect Africa Scholarship Award, H3ABioNet/SANBio Scholarship and North-West University for offering me bursary/scholarship award to pursue the PhD degree. -
Nitroxide-Mediated Polymerization
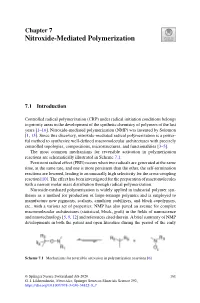
Chapter 7 Nitroxide-Mediated Polymerization 7.1 Introduction Controlled radical polymerization (CRP) under radical initiation conditions belongs to priority areas in the development of the synthetic chemistry of polymers of the last years [1–16]. Nitroxide-mediated polymerization (NMP) was invented by Solomon [1, 13]. Since this discovery, nitroxide-mediated radical polymerization is a power- ful method to synthesize well-defined macromolecular architectures with precisely controlled topologies, compositions, microstructures, and functionalities [3–5]. The most common mechanisms for reversible activation in polymerization reactions are schematically illustrated in Scheme 7.1. Persistent radical effect (PRE) occurs when two radicals are generated at the same time, at the same rate, and one is more persistent than the other, the self-termination reactions are lowered, leading to an unusually high selectivity for the cross-coupling reaction [10]. The effect has been investigated for the preparation of macromolecules with a narrow molar mass distribution through radical polymerization. Nitroxide-mediated polymerization is widely applied in industrial polymer syn- theses as a method for production of large-tonnage polymers and is employed to manufacture new pigments, sealants, emulsion stabilizers, and block copolymers, etc., with a various set of properties. NMP has also paved an avenue for complex macromolecular architectures (statistical, block, graft) in the fields of nanoscience and nanotechnology [5, 9, 12] and references cited therein. A brief summary of NMP developments in both the patent and open literature during the period of the early Scheme 7.1 Mechanisms for reversible activation in polymerization reactions [6] © Springer Nature Switzerland AG 2020 161 G. I. Likhtenshtein, Nitroxides, Springer Series in Materials Science 292, https://doi.org/10.1007/978-3-030-34822-9_7 162 7 Nitroxide-Mediated Polymerization 1980–2000 was presented in [11]. -

Exosomes Confer Chemoresistance to Pancreatic Cancer Cells By
FULL PAPER British Journal of Cancer (2017) 116, 609–619 | doi: 10.1038/bjc.2017.18 Keywords: chemoresistance; exosomes; pancreatic cancer; ROS; microRNA Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK Girijesh Kumar Patel1, Mohammad Aslam Khan1, Arun Bhardwaj1, Sanjeev K Srivastava1, Haseeb Zubair1, Mary C Patton1, Seema Singh1,2, Moh’d Khushman3 and Ajay P Singh*,1,2 1Department of Oncologic Sciences, Mitchell Cancer Institute, University of South Alabama, Mobile, AL, USA; 2Department of Biochemistry and Molecular Biology, College of Medicine, University of South Alabama, Mobile, AL, USA and 3Department of Interdisciplinary Clinical Oncology, Mitchell Cancer Institute, University of South Alabama, Mobile, AL, USA Background: Chemoresistance is a significant clinical problem in pancreatic cancer (PC) and underlying molecular mechanisms still remain to be completely understood. Here we report a novel exosome-mediated mechanism of drug-induced acquired chemoresistance in PC cells. Methods: Differential ultracentrifugation was performed to isolate extracellular vesicles (EVs) based on their size from vehicle- or gemcitabine-treated PC cells. Extracellular vesicles size and subtypes were determined by dynamic light scattering and marker profiling, respectively. Gene expression was examined by qRT-PCR and/or immunoblot analyses, and direct targeting of DCK by miR-155 was confirmed by dual-luciferase 30-UTR reporter assay. Flow cytometry was performed to examine the apoptosis indices and reactive oxygen species (ROS) levels in PC cells using specific dyes. Cell viability was determined using the WST-1 assay. Results: Conditioned media (CM) from gemcitabine-treated PC cells (Gem-CM) provided significant chemoprotection to subsequent gemcitabine toxicity and most of the chemoresistance conferred by Gem-CM resulted from its EVs fraction. -

Integrative Systems Biology– Renal Diseases: a Road to a Holist View of Chronic Disease Mechanism
Integrative Systems biology– Renal Diseases: A road to a holist view of chronic disease mechanism Matthias Kretzler Div. Nephrology / Internal Medicine Computational Medicine and Bioinformatics University of Michigan Medical School The challenge in chronic disease • Descriptive disease categorization with multiple pathogenetic mechanisms § Problems of ‘mixed bag’ diseases: • Unpredictable disease course and response to therapy • Nephrology as an ‘art of trial and error’ • Shift in our disease paradigms: § Mechanism based patient management • Define the disease process active in the individual patient – Base prognosis on specific disease process – Target therapy to interfere with the mechanism currently destroying endorgan function Molecular Nephrology approach Clinical outcome research Genetics Molecular Pathology Molecular Epigenetics Phenotyping Genomics Functional Clinical research Disease Genomics Biobanks Proteomics Model systems Animal models Molecular interaction In vitro tissue culture model systems Organ culture and development Tower of Babylon: Search for the universal language for the medicine of the 21st century Pieter Bruegl: 1563. Kunsthistorisches Museum Wien Molecular Nephrology approach Clinical outcome research Genetics Molecular Pathology Molecular Epigenetics Phenotyping Genomics Functional Clinical research Disease Genomics Integrative Biobanks Proteomics Biology (Physiology) Model systems Animal models Molecular interaction of renal disease In vitro tissue culture model systems Organ culture and development Systems -

Ribonucleotide Reductase Subunit M2B Deficiency Leads To
Am J Transl Res 2018;10(11):3635-3649 www.ajtr.org /ISSN:1943-8141/AJTR0084684 Original Article Ribonucleotide reductase subunit M2B deficiency leads to mitochondrial permeability transition pore opening and is associated with aggressive clinicopathologic manifestations of breast cancer Lijun Xue1*, Xiyong Liu2,6*, Qinchuan Wang3,4, Charlie Q Liu3, Yunru Chen3, Wei Jia5, Ronhong Hsie6, Yifan Chen8, Frank Luh2,6, Shu Zheng7, Yun Yen2,6,8 1Department of Pathology, Loma Linda University Medical Center, Loma Linda, CA 92354, USA; 2Sino-American Cancer Foundation, California Cancer Institute, Temple, CA 91780, USA; 3Department of Molecular Pharmacology, Beckman Research Institute, City of Hope Comprehensive Cancer Center, Duarte, CA 91010, USA; 4Surgical Oncology, Sir Runrun Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China; 5Cancer Epidemiology Program, University of Hawaii Cancer Center, Honolulu, HI 96813, USA; 6TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei, Taiwan, ROC; 7Cancer Institute, Zhejiang University, Hangzhou 310009, Zhejiang, China; 8PhD Program of Cancer Biology and Drug Discovery, Taipei Medical University, Taipei, Taiwan, ROC. *Co-first authors. Received August 27, 2018; Accepted October 19, 2018; Epub November 15, 2018; Published November 30, 2018 Abstract: Ribonucleotide reductase small subunit M2B (RRM2B) plays an essential role in maintaining mitochon- drial homeostasis. Mitochondrial permeability transition pore (MPTP) is a key regulator of mitochondrial homeo- stasis. MPTP contributes to cell death and is crucial in cancer progression. RRM2B’s relation to MPTP is not well known, and the role of RRM2B in cancer progression is controversial. Here, our aim was to study the role of RRM2B in regulating MPTP and the association between RRM2B and clinicopathological manifestations in breast cancer. -

Knockdown of RRM1 with Adenoviral Shrna Vectors to Inhibit Tumor Cell Viability and Increase Chemotherapeutic Sensitivity to Gemcitabine in Bladder Cancer Cells
International Journal of Molecular Sciences Article Knockdown of RRM1 with Adenoviral shRNA Vectors to Inhibit Tumor Cell Viability and Increase Chemotherapeutic Sensitivity to Gemcitabine in Bladder Cancer Cells Xia Zhang 1, Rikiya Taoka 1,*, Dage Liu 2, Yuki Matsuoka 1, Yoichiro Tohi 1 , Yoshiyuki Kakehi 1 and Mikio Sugimoto 1 1 Department of Urology, Faculty of Medicine, Kagawa University, 1750-1 Ikenobe, Miki-cho, Kita-gun, Kagawa 761-0793, Japan; [email protected] (X.Z.); [email protected] (Y.M.); [email protected] (Y.T.); [email protected] (Y.K.); [email protected] (M.S.) 2 Department of General Thoracic Surgery, Faculty of Medicine, Kagawa University, 1750-1 Ikenobe, Miki-cho, Kita-gun, Kagawa 761-0793, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-87-891-2202 Abstract: RRM1—an important DNA replication/repair enzyme—is the primary molecular gem- citabine (GEM) target. High RRM1-expression associates with gemcitabine-resistance in various cancers and RRM1 inhibition may provide novel cancer treatment approaches. Our study eluci- dates how RRM1 inhibition affects cancer cell proliferation and influences gemcitabine-resistant bladder cancer cells. Of nine bladder cancer cell lines investigated, two RRM1 highly expressed cells, 253J and RT112, were selected for further experimentation. An RRM1-targeting shRNA was Citation: Zhang, X.; Taoka, R.; Liu, cloned into adenoviral vector, Ad-shRRM1. Gene and protein expression were investigated using D.; Matsuoka, Y.; Tohi, Y.; Kakehi, Y.; real-time PCR and western blotting. -

Mitochondrial Hepatopathies Etiology and Genetics the Hepatocyte Mitochondrion Can Function Both As a Cause and As a Target of Liver Injury
Mitochondrial Hepatopathies Etiology and Genetics The hepatocyte mitochondrion can function both as a cause and as a target of liver injury. Most mitochondrial hepatopathies involve defects in the mitochondrial respiratory chain enzyme complexes (Figure 1). Resultant dysfunction of mitochondria yields deficient oxidative phosphorylation (OXPHOS), increased generation of reactive oxygen species (ROS), accumulation of hepatocyte lipid, impairment of other metabolic pathways and activation of both apoptotic and necrotic pathways of cellular death. Figure 1: Since the mitochondria are under dual control of nuclear DNA and mitochondrial DNA (mtDNA), mutations in genes of both classes have been associated with inherited mitochondrial myopathies, encephalopathies, and hepatopathies. Autosomal nuclear gene defects affect a variety of mitochondrial processes such as protein assembly, mtDNA synthesis and replication (e.g., deoxyguanosine kinase [dGUOK]) and DNA polymerase gamma [POLG]), and transport of nucleotides or metals. MPV17 (function unknown) and RRM2B (encoding the cytosolic p53-inducible ribonucleotide reductase small subunit) are two genes recently identified as also causing mtDNA depletion syndrome and liver failure, as has TWINKLE, TRMU, and SUCLG1. Most children with mitochondrial hepatopathies have identified or presumed mutations in these nuclear genes, rather than mtDNA genes. A classification of primary mitochondrial hepatopathies involving energy metabolism is presented in Table 1. Drug interference with mtDNA replication is now recognized as a cause of acquired mtDNA depletion that can result in liver failure, lactic acidosis, and myopathy in human immunodeficiency virus infected patients and, previously, in hepatitis B virus patients treated with nucleoside reverse transcriptase inhibitors. Current estimates suggest a minimum prevalence of all mitochondrial diseases of 11.5 cases per 100,000 individuals, or 1 in 8500 of the general population. -

Tremblay Robinlee.Pdf
Expression and Characterisation ofa Gene Enc oding RbpD, an RNA- Bind ing Protein in Anabaena sp. strain PeC 7120 by Rob in lee Tremblay A lhesis submitted to the Scltool of Graduale Studies in partial fulfilment of the requirements fOl" the degree of Master of Science Department of BiochemistrylFacultyof Science Memorial University of Newfoundland January 2000 SI.JOM'S Newfoun dland Abs t ra ct The RNA-binding protein RbpD, from the cyano bacterium Anaba ena sp, strain Pe C 7120 was expressed in £Sch~ ric h ia coli and successfully purified using me IMPACT I system (New England Biolabs). The rbp D gene was cloned into the pCYBt expre ssion vector by using polymerase chain reaction to introduce Ndel and SapI restriction sites at the 5' end 3' ends of the gene respect ively. The 3'.-end mutagenesis also chan ged the stop codon into a cysteine codon. The resulting gene encoded a fusion protein consisting of RbpD, the Saccharomyces cerev isiae VMA intein and a chitin binding domain.. Expressi on of the fusion protein was observed in £ coli strain MCI061 but Western blot analysis using an intein-directed ant ibody indicated that significant in vivo fmeln-direcred splicing of the fusion protein occurred. We were unable to eliminate this problem; no fusion protein expression was observed in 8 other E coli strains tested. Wild -type RbpD was purified following binding of the fusion protein 10 a chitin column and overnight cleavage in the presence of a reducing agent, dlthicthrehc l. A number of modifications to the manufacturer' s purification protocol were found to be necessary for success ful purification. -

Purification and Characterisation of a Protease (Tamarillin) from Tamarillo Fruit
Purification and characterisation of a protease (tamarillin) from tamarillo fruit Item Type Article Authors Li, Zhao; Scott, Ken; Hemar, Yacine; Zhang, Huoming; Otter, Don Citation Li Z, Scott K, Hemar Y, Zhang H, Otter D (2018) Purification and characterisation of a protease (tamarillin) from tamarillo fruit. Food Chemistry. Available: http://dx.doi.org/10.1016/ j.foodchem.2018.02.091. Eprint version Post-print DOI 10.1016/j.foodchem.2018.02.091 Publisher Elsevier BV Journal Food Chemistry Rights NOTICE: this is the author’s version of a work that was accepted for publication in Food Chemistry. Changes resulting from the publishing process, such as peer review, editing, corrections, structural formatting, and other quality control mechanisms may not be reflected in this document. Changes may have been made to this work since it was submitted for publication. A definitive version was subsequently published in Food Chemistry, [, , (2018-02-16)] DOI: 10.1016/j.foodchem.2018.02.091 . © 2018. This manuscript version is made available under the CC-BY-NC-ND 4.0 license http://creativecommons.org/licenses/by-nc-nd/4.0/ Download date 29/09/2021 23:19:14 Link to Item http://hdl.handle.net/10754/627180 Accepted Manuscript Purification and characterisation of a protease (tamarillin) from tamarillo fruit Zhao Li, Ken Scott, Yacine Hemar, Huoming Zhang, Don Otter PII: S0308-8146(18)30327-3 DOI: https://doi.org/10.1016/j.foodchem.2018.02.091 Reference: FOCH 22475 To appear in: Food Chemistry Received Date: 25 October 2017 Revised Date: 13 February 2018 Accepted Date: 16 February 2018 Please cite this article as: Li, Z., Scott, K., Hemar, Y., Zhang, H., Otter, D., Purification and characterisation of a protease (tamarillin) from tamarillo fruit, Food Chemistry (2018), doi: https://doi.org/10.1016/j.foodchem. -

Arsenic Hexoxide Has Differential Effects on Cell Proliferation And
www.nature.com/scientificreports OPEN Arsenic hexoxide has diferential efects on cell proliferation and genome‑wide gene expression in human primary mammary epithelial and MCF7 cells Donguk Kim1,7, Na Yeon Park2,7, Keunsoo Kang3, Stuart K. Calderwood4, Dong‑Hyung Cho2, Ill Ju Bae5* & Heeyoun Bunch1,6* Arsenic is reportedly a biphasic inorganic compound for its toxicity and anticancer efects in humans. Recent studies have shown that certain arsenic compounds including arsenic hexoxide (AS4O6; hereafter, AS6) induce programmed cell death and cell cycle arrest in human cancer cells and murine cancer models. However, the mechanisms by which AS6 suppresses cancer cells are incompletely understood. In this study, we report the mechanisms of AS6 through transcriptome analyses. In particular, the cytotoxicity and global gene expression regulation by AS6 were compared in human normal and cancer breast epithelial cells. Using RNA‑sequencing and bioinformatics analyses, diferentially expressed genes in signifcantly afected biological pathways in these cell types were validated by real‑time quantitative polymerase chain reaction and immunoblotting assays. Our data show markedly diferential efects of AS6 on cytotoxicity and gene expression in human mammary epithelial normal cells (HUMEC) and Michigan Cancer Foundation 7 (MCF7), a human mammary epithelial cancer cell line. AS6 selectively arrests cell growth and induces cell death in MCF7 cells without afecting the growth of HUMEC in a dose‑dependent manner. AS6 alters the transcription of a large number of genes in MCF7 cells, but much fewer genes in HUMEC. Importantly, we found that the cell proliferation, cell cycle, and DNA repair pathways are signifcantly suppressed whereas cellular stress response and apoptotic pathways increase in AS6‑treated MCF7 cells. -

A Ferrous-Triapine Complex Mediates Formation of Reactive Oxygen Species That Inactivate Human Ribonucleotide Reductase
586 A Ferrous-triapine complex mediates formation of reactive oxygen species that inactivate human ribonucleotide reductase Jimin Shao,1,2 Bingsen Zhou,1 Angel J. Di Bilio,3 cytotoxicity assays. These results indicate that Triapine- Lijun Zhu,1,2 Tieli Wang,1 Christina Qi,1 induced inhibition of ribonucleotide reductase is caused by Jennifer Shih,1 and Yun Yen1 ROS. We suggest that ROS may ultimately be responsible for the pharmacologic effects of Triapine in vivo. [Mol 1Department of Medical Oncology and Therapeutic Research, Cancer Ther 2006;5(3):586–92] City of Hope National Medical Center, Duarte, California; 2Department of Basic Medical Sciences, Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of Introduction 3 China; Department of Chemistry, California Institute of Ribonucleotide reductases catalyze the reduction of Technology, Pasadena, California ribonucleotides to deoxyribonucleotides, which are re- quired for DNA replication and repair (1, 2). Human Abstract ribonucleotide reductase is a protein tetramer featuring Ribonucleotide reductase plays a central role in cell two identical large (hRRM1) and two identical small proliferation by supplying deoxyribonucleotide precursors (hRRM2 or p53R2) subunits. hRRM1 harbors the sub- for DNA synthesis and repair. The holoenzyme is a protein strate-catalytic and allosteric regulation sites. The small tetramer that features two large (hRRM1) and two small subunit contains an oxygen-linked diferric cluster and a (hRRM2 or p53R2) subunits. The small subunit contains a stable tyrosyl radical that are needed for function (Fig. 1A; refs. 1–4). p53R2 is a newly identified p53-inducible di-iron cluster/tyrosyl radical cofactor that is essential for f enzyme activity. -

TRACE: Tennessee Research and Creative Exchange
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 8-2009 Structure-Function Studies of the Large Subunit of Ribonucleotide Reductase from Homo sapiens and Saccharomyces cerevisiae James Wesley Fairman University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Biochemistry, Biophysics, and Structural Biology Commons Recommended Citation Fairman, James Wesley, "Structure-Function Studies of the Large Subunit of Ribonucleotide Reductase from Homo sapiens and Saccharomyces cerevisiae. " PhD diss., University of Tennessee, 2009. https://trace.tennessee.edu/utk_graddiss/49 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by James Wesley Fairman entitled "Structure- Function Studies of the Large Subunit of Ribonucleotide Reductase from Homo sapiens and Saccharomyces cerevisiae." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Doctor of Philosophy, with a major in Biochemistry and Cellular and Molecular Biology. Chris G. Dealwis,