Computational Drug Repurposing Study Elucidating Simultaneous Inhibition of Entry and Replication of Novel Corona Virus by Grazoprevir
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Development of Antiviral Agents for Enteroviruses
Journal of Antimicrobial Chemotherapy (2008) 62, 1169–1173 doi:10.1093/jac/dkn424 Advance Access publication 18 October 2008 Development of antiviral agents for enteroviruses Tzu-Chun Chen1–3, Kuo-Feng Weng1,2, Shih-Cheng Chang1,2, Jing-Yi Lin1,2, Peng-Nien Huang1,2 and Shin-Ru Shih1,2,4,5* 1Research Center for Emerging Viral Infections, Chang Gung University, Taoyuan, Taiwan; 2Department of Medical Biotechnology and Laboratory Science, Chang Gung University, Taoyuan, Taiwan; 3Institute Downloaded from https://academic.oup.com/jac/article/62/6/1169/774341 by guest on 26 September 2021 of Molecular Medicine, National Tsing Hua University, Hsinchu, Taiwan; 4Clinical Virology Laboratory, Chang Gung Memorial Hospital, Taoyuan, Taiwan; 5Division of Biotechnology and Pharmaceutical Research, National Health Research Institutes, Chunan, Taiwan Enteroviruses (EVs) are common human pathogens that are associated with numerous disease symptoms in many organ systems of the body. Although EV infections commonly cause mild or non- symptomatic illness, some of them are associated with severe diseases such as CNS complications. The current absence of effective vaccines for most viral infection and no available antiviral drugs for the treatment of EVs highlight the urgency and significance of developing antiviral agents. Several key steps in the viral life cycle are potential targets for blocking viral replication. This article reviews recent studies of antiviral developments for EVs based on various molecular targets that interrupt viral attach- ment, viral translation, polyprotein processing and RNA replication. Keywords: capsid proteins, viral proteases, viral RNA replication, 50 untranslated region Introduction EVs such as echovirus 6, 9 and 30 and CVB5 were the most common causes of aseptic meningitis in children.4 EV70 and Enteroviruses (EVs) are a common cause of infections in CVA24 were associated with acute haemorrhagic conjunctivitis.4 humans, especially children. -

The Treatment of Herpes Simplex Virus Epithelial Keratitis
THE TREATMENT OF HERPES SIMPLEX VIRUS EPITHELIAL KERATITIS BY Kirk R. Wilhelmus, MD, MPH ABSTRACT Purpose: Epithelial keratitis is the most common presentation of ocular infection by herpes simplex virus (HSV). Quantitative assessment of available therapy is needed to guide evidence-based ophthalmology. This study aimed to compare the efficacy of various treatments for dendritic or geographic HSV epithelial keratitis and to evaluate the role of various clinical characteristics on epithelial healing. Methods: Following a systematic review of the literature, information from clinical trials of HSV dendritic or geographic epithelial keratitis was extracted, and the methodological quality of each study was scored. Methods of epithelial cau- terization and curettage were grouped as relatively equivalent physicochemical therapy, and solution and ointment for- mulations of a given topical antiviral agent were combined. The proportion healed with 1 week of therapy, a scheduled follow-up day that approximated the average time of resolution with antiviral therapy, was selected as the primary out- come based on a masked evaluation of maximum treatment differences in published healing curves. The proportion healed at 14 days was recorded as supplemental information. Fixed-effects and random-effects meta-analysis models were used to obtain summary estimates by pooling results from comparative treatment trials. Hypotheses about which prognostic factors might affect epithelial healing during antiviral therapy were developed by multivariate analysis of the Herpetic Eye Disease Study dataset. Results: After excluding 48 duplicate reports, 14 nonrandomized studies, 15 studies with outdated or similar treatments, and 29 trials lacking sufficient data on healing or accessibility, 76 primary reports were identified. -
(12) United States Patent (10) Patent No.: US 8,993,581 B2 Perrine Et Al
US00899.3581B2 (12) United States Patent (10) Patent No.: US 8,993,581 B2 Perrine et al. (45) Date of Patent: Mar. 31, 2015 (54) METHODS FOR TREATINGVIRAL (58) Field of Classification Search DSORDERS CPC ... A61K 31/00; A61K 31/166; A61K 31/185: A61K 31/233; A61K 31/522: A61K 38/12: (71) Applicant: Trustees of Boston University, Boston, A61K 38/15: A61K 45/06 MA (US) USPC ........... 514/263.38, 21.1, 557, 565, 575, 617; 424/2011 (72) Inventors: Susan Perrine, Weston, MA (US); Douglas Faller, Weston, MA (US) See application file for complete search history. (73) Assignee: Trustees of Boston University, Boston, (56) References Cited MA (US) U.S. PATENT DOCUMENTS (*) Notice: Subject to any disclaimer, the term of this 3,471,513 A 10, 1969 Chinn et al. patent is extended or adjusted under 35 3,904,612 A 9/1975 Nagasawa et al. U.S.C. 154(b) by 0 days. (Continued) (21) Appl. No.: 13/915,092 FOREIGN PATENT DOCUMENTS (22) Filed: Jun. 11, 2013 CA 1209037 A 8, 1986 CA 2303268 A1 4f1995 (65) Prior Publication Data (Continued) US 2014/OO45774 A1 Feb. 13, 2014 OTHER PUBLICATIONS Related U.S. Application Data (63) Continuation of application No. 12/890,042, filed on PCT/US 10/59584 Search Report and Written Opinion mailed Feb. Sep. 24, 2010, now abandoned. 11, 2011. (Continued) (60) Provisional application No. 61/245,529, filed on Sep. 24, 2009, provisional application No. 61/295,663, filed on Jan. 15, 2010. Primary Examiner — Savitha Rao (74) Attorney, Agent, or Firm — Nixon Peabody LLP (51) Int. -

And Ritonavir-Boosted HIV Protease Inhibitors
16 February 2012 EMA/CHMP/117973/2012 EMEA/H/C/002332/II/0004 Questions and answers on drug interactions between Victrelis (boceprevir) and ritonavir-boosted HIV protease inhibitors The European Medicines Agency has recommended changes to the prescribing information for Victrelis (boceprevir), a medicine used to treat hepatitis C, after a drug interaction study identified interactions between Victrelis and medicines used to treat HIV called ritonavir-boosted HIV protease inhibitors. These interactions could potentially reduce the effectiveness of these medicines if used together in patients being treated for both hepatitis C and HIV. The Agency’s Committee for Medicinal Products for Human Use (CHMP) has recommended the changes to ensure doctors are informed of these interactions while further data are awaited to assess the clinical impact of these drug interaction findings on these patients. What is Victrelis? Victrelis is a medicine used to treat long-term hepatitis C genotype 1 (a disease of the liver due to infection with the hepatitis C virus) in adults with compensated liver disease who have not been treated before or whose previous treatment has failed. Compensated liver disease is when the liver is damaged but is still able to work normally. Victrelis is given in combination with two other medicines, peginterferon alfa and ribavirin. The active substance in Victrelis, boceprevir, is a protease inhibitor which blocks an enzyme called HCV NS3 protease found on the hepatitis C genotype 1 virus. Victrelis was authorised in the EU in July 2011. What is the issue with Victrelis? In January 2012, the EMA was informed of the results of a study in healthy volunteers which identified drug interactions between Victrelis and the antiviral medicines atazanavir, darunavir and lopinavir, which are used to treat HIV. -

Telaprevir (Incivek)
© Hepatitis C Online PDF created September 25, 2021, 4:19 pm Telaprevir (Incivek) Discontinued. This treatment has been discontinued. Table of Contents Telaprevir Incivek Summary Drug Summary Adverse Effects Class and Mechanism Manufacturer for United States FDA Status Indications Dosing Clinical Use Cost and Medication Access Resistance Key Drug Interactions Full Prescribing Information Figures Drug Summary Although telaprevir was a promising direct-acting antiviral agent that had impact in the hepatitis C treatment field during 2011 to 2013, it was subsequently replaced by newer direct-acting antiviral agents that were more effective, better tolerated, and more convenient. Based on the dwindling role of telaprevir after newer direct-acting antiviral agents were approved, Vertex pharmaceuticals discontinued the sales and distribution of telaprevir in the United States in October 2014. Telaprevir does have some current importance since persons who previously failed a telaprevir-based regimen may have developed resistant associated variants, which could potentially impact subsequent therapy. Adverse Effects The most significant adverse effects reported in the main registration trials and in post-marketing experience were rash, anorectal complaints, and anemia. When comparing triple therapy of telaprevir, peginterferon, and ribavirin with dual therapy of peginterferon and ribavirin alone significant differences were noted with rash (56% versus 34%), anemia (36% versus 17%), and anorectal complaints that include anorectal discomfort, anal pruritus, and hemorrhoids (29% versus 7%). In most cases, the rash that develops is eczematous or maculopapular in character and mild to moderate in severity; the rash is typically manageable with good skin care and topical emollients or corticosteroids. In some instances, however, telaprevir has caused serious skin Page 1/5 rashes, including Steven's Johnson Syndrome (SJS), Drug Reaction with Eosinophilia and Systemic Symptoms (DRESS), and Toxic Epidermal Necrolysis (TEN). -
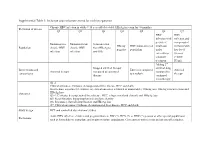
Inclusion and Exclusion Criteria for Each Key Question
Supplemental Table 1: Inclusion and exclusion criteria for each key question Chronic HBV infection in adults ≥ 18 year old (detectable HBsAg in serum for >6 months) Definition of disease Q1 Q2 Q3 Q4 Q5 Q6 Q7 HBV HBV infection with infection and persistent compensated Immunoactive Immunotolerant Seroconverted HBeAg HBV mono-infected viral load cirrhosis with Population chronic HBV chronic HBV from HBeAg to negative population under low level infection infection anti-HBe entecavir or viremia tenofovir (<2000 treatment IU/ml) Adding 2nd Stopped antiviral therapy antiviral drug Interventions and Entecavir compared Antiviral Antiviral therapy compared to continued compared to comparisons to tenofovir therapy therapy continued monotherapy Q1-2: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Intermediate outcomes (if evidence on clinical outcomes is limited or unavailable): HBsAg loss, HBeAg seroconversion and Outcomes HBeAg loss Q3-4: Cirrhosis, decompensated liver disease, HCC, relapse (viral and clinical) and HBsAg loss Q5: Renal function, hypophosphatemia and bone density Q6: Resistance, flare/decompensation and HBeAg loss Q7: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Study design RCT and controlled observational studies Acute HBV infection, children and pregnant women, HIV (+), HCV (+) or HDV (+) persons or other special populations Exclusions such as hemodialysis, transplant, and treatment failure populations. Co treatment with steroids and uncontrolled studies. Supplemental Table 2: Detailed Search Strategy: Ovid Database(s): Embase 1988 to 2014 Week 37, Ovid MEDLINE(R) In-Process & Other Non- Indexed Citations and Ovid MEDLINE(R) 1946 to Present, EBM Reviews - Cochrane Central Register of Controlled Trials August 2014, EBM Reviews - Cochrane Database of Systematic Reviews 2005 to July 2014 Search Strategy: # Searches Results 1 exp Hepatitis B/dt 26410 ("hepatitis B" or "serum hepatitis" or "hippie hepatitis" or "injection hepatitis" or 2 178548 "hepatitis type B").mp. -

The Evolution of Pleconaril: Modified O-Alkyl Linker Analogs Have
molecules Communication The Evolution of Pleconaril: Modified O-Alkyl Linker Analogs Have Biological Activity towards Coxsackievirus B3 Nancy 1, 2, 1 3 Alexandrina Volobueva y, Anna Egorova y, Anastasia Galochkina , Sean Ekins , Vladimir Zarubaev 1 and Vadim Makarov 2,* 1 Saint-Petersburg Pasteur Institute, Mira str., 14, 197101 Saint Petersburg, Russia; [email protected] (A.V.); [email protected] (A.G.); [email protected] (V.Z.) 2 Bach Institute of Biochemistry, Research Center of Biotechnology of the Russian Academy of Sciences, Leninsky prospect, 33, build. 2, 119071 Moscow, Russia; [email protected] 3 Collaborations Pharmaceuticals, Inc., 840 Main Campus Drive, Lab 3510, Raleigh, NC 27606, USA; [email protected] * Correspondence: [email protected] These authors contributed equally to this work. y Received: 10 February 2020; Accepted: 13 March 2020; Published: 16 March 2020 Abstract: Coxsackieviruses type B are one of the most common causes of mild upper respiratory and gastrointestinal illnesses. At the time of writing, there are no approved drugs for effective antiviral treatment for Coxsackieviruses type B. We used the core-structure of pleconaril, a well-known antienteroviral drug candidate, for the synthesis of novel compounds with O-propyl linker modifications. Some original compounds with 4 different linker patterns, such as sulfur atom, ester, amide, and piperazine, were synthesized according to five synthetic schemes. The cytotoxicity and bioactivity of 14 target compounds towards Coxsackievirus B3 Nancy were examined. Based on the results, the values of 50% cytotoxic dose (CC50), 50% virus-inhibiting dose (IC50), and selectivity index (SI) were calculated for each compound. Several of the novel synthesized derivatives exhibited a strong anti-CVB3 activity (SI > 20 to > 200). -

Which Drugs Are Most Effective for Recurrent Herpes Labialis?
Evidence-based answers from the clinical inquiries Family Physicians Inquiries Network Eiko Tubridy, MD; Gary Kelsberg, MD Valley Family Residency Which drugs are most Program, Renton, Wash Leilani St Anna, MLIS, effective for recurrent AHIP University of Washington Health Sciences Libraries, herpes labialis? Seattle AssistanT EDITOR EvidEncE-basEd answEr Jon O. neher, MD Valley Family Residency daily oral acyclovir or vala- ing to the agent used: valacyclovir reduces Program, Renton, Wash A cyclovir may help prevent her- both healing time and duration of pain, pes simplex labialis (HSL) recurrences famciclovir reduces both in one dosage (strength of recommendation [SOR]: B, form but not another, and acyclovir reduces meta-analysis of randomized controlled only pain duration (SOR: B, single RCTs). trials [RCTs] with heterogeneous results). Several topical medications (acyclovir, No trials compare oral or topical treat- penciclovir, docosanol) modestly decrease ments for HSL outbreaks against each oth- healing time and pain duration—typically er. Oral antivirals modestly reduce healing by less than a day—and require multiple time and duration of pain, varying accord- doses per day (SOR: B, multiple RCTs). Evidence summary The authors of the meta-analysis noted A systematic review and meta-analysis of the that although 9 studies favored the use of an effectiveness of oral and topical nucleoside antiviral drug, only 4 showed statistically sig- antiviral agents to prevent recurrent HSL in nificant differences when compared with pla- immunocompetent people found 11 RCTs cebo, and none of them had a low risk of bias. with a total of 1250 patients that compared They concluded that the review supported us- an active drug against placebo.1 The medi- ing oral acyclovir and valacyclovir to prevent cations were topical 5% acyclovir, topical 1% recurrent HSL.1 penciclovir, and oral acyclovir, valacyclovir, or famciclovir in various doses. -

Telaprevir for HIV/Hepatitis C Virus–Coinfected Patients Failing
HIV/AIDS MAJOR ARTICLE Telaprevir for HIV/Hepatitis C Virus–Coinfected Patients Failing Treatment With Pegylated Interferon/Ribavirin (ANRS HC26 TelapreVIH): An Open-Label, Single-Arm, Phase 2 Trial Downloaded from https://academic.oup.com/cid/article/59/12/1768/2895305 by guest on 01 October 2021 Laurent Cotte,1 Joséphine Braun,2 Caroline Lascoux-Combe,3 Corine Vincent,2 Marc-Antoine Valantin,4 Philippe Sogni,5 Karine Lacombe,6 Didier Neau,7 Hugues Aumaitre,8 Dominique Batisse,9 Pierre de Truchis,10 Anne Gervais,11 Christian Michelet,12 Philippe Morlat,13 Daniel Vittecoq,14 Isabelle Rosa,15 Inga Bertucci,16 Stéphane Chevaliez,17 Jean-Pierre Aboulker,2 and Jean-Michel Molina3; for the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) HC26 Study Groupa 1Hospices Civils de Lyon, Croix-Rousse Hospital, and INSERM U1052, 2INSERM SC10-US019, Villejuif, 3Assistance Publique–Hôpitaux de Paris (AP-HP), Saint-Louis Hospital, University of Paris VII Denis Diderot, and Sorbonne Paris-Cité, INSERM U941, 4AP-HP, Pitié-Salpêtrière Hospital, and UMR-S 943, INSERM, 5AP-HP, Cochin Hospital and Paris Descartes University, INSERM U-1016, 6AP-HP, Saint-Antoine Hospital, Sorbonne Universités, UPMC University Paris 06, UMR-S1136, 7Pellegrin University Hospital, Bordeaux, 8Saint-Jean Hospital, Perpignan, 9AP-HP, Georges Pompidou European Hospital, Paris, 10AP-HP, Raymond Poincaré Hospital, Garches, 11AP-HP, Bichat-Claude Bernard Hospital, Paris, 12Pontchaillou University Hospital, Rennes, 13Saint-André University Hospital, Bordeaux, 14AP-HP, Bicètre Hospital, Le Kremlin-Bicètre, 15Créteil Hospital, 16French National Agency for Research on AIDS and Viral Hepatitis, Paris, and 17AP-HP, Henri Mondor Hospital, Créteil, France (See the Editorial Commentary by Rockstroh on pages 1777–8.) Background. -

Review of Sezary Syndrome
REVIEWS Sezary syndrome: Immunopathogenesis, literature review of therapeutic options, and recommendations for therapy by the United States Cutaneous Lymphoma Consortium (USCLC) EliseA.Olsen,MD,a Alain H. Rook, MD,b John Zic, MD,c Youn Kim, MD,d PierluigiPorcu,MD,e Christiane Querfeld, MD,f Gary Wood, MD,g Marie-France Demierre, MD,h Mark Pittelkow, MD,i Lynn D. Wilson, MD, MPH,j Lauren Pinter-Brown, MD,k Ranjana Advani, MD,d Sareeta Parker, MD,l Ellen J. Kim, MD,b Jacqueline M. Junkins-Hopkins, MD,m Francine Foss, MD,j Patrick Cacchio, BS,a and Madeleine Duvic, MDn Durham, North Carolina; Philadelphia, Pennsylvania; Nashville, Tennessee; Palo Alto and Los Angeles, California; Columbus, Ohio; Chicago, Illinois;Madison,Wisconsin;Boston,Massachusetts; Rochester, Minnesota; New Haven, Connecticut; Atlanta, Georgia; Baltimore, Maryland; and Houston, Texas Sezary syndrome (SS) has a poor prognosis and few guidelines for optimizing therapy. The US Cutaneous Lymphoma Consortium, to improve clinical care of patients with SS and encourage controlled clinical trials of promising treatments, undertook a review of the published literature on therapeutic options for SS. An overview of the immunopathogenesis and standardized review of potential current treatment options for SS including metabolism, mechanism of action, overall efficacy in mycosis fungoides and SS, and common or concerning adverse effects is first discussed. The specific efficacy of each treatment for SS, both as monotherapy and combination therapy, is then reported using standardized criteria for both SS and response to therapy with the type of study defined by a modification of the US Preventive Services guidelines for evidence-based medicine. -

Multipurpose Tenofovir Disoproxil Fumarate Electrospun Fibers for the Prevention of HIV-1 and HSV-2 Infections
University of Louisville ThinkIR: The University of Louisville's Institutional Repository Electronic Theses and Dissertations 8-2016 Multipurpose tenofovir disoproxil fumarate electrospun fibers for the prevention of HIV-1 and HSV-2 infections. Kevin Tyo Follow this and additional works at: https://ir.library.louisville.edu/etd Part of the Other Chemicals and Drugs Commons, Polymer Chemistry Commons, Preventive Medicine Commons, and the Virus Diseases Commons Recommended Citation Tyo, Kevin, "Multipurpose tenofovir disoproxil fumarate electrospun fibers for the prevention of HIV-1 and HSV-2 infections." (2016). Electronic Theses and Dissertations. Paper 2486. https://doi.org/10.18297/etd/2486 This Master's Thesis is brought to you for free and open access by ThinkIR: The University of Louisville's Institutional Repository. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of ThinkIR: The University of Louisville's Institutional Repository. This title appears here courtesy of the author, who has retained all other copyrights. For more information, please contact [email protected]. MULTIPURPOSE TENOFOVIR DISOPROXIL FUMARATE ELECTROSPUN FIBERS FOR THE PREVENTION OF HIV-1 AND HSV-2 INFECTIONS By Kevin Tyo B.S. Virginia Tech, 2010 A Thesis Submitted to the Faculty of the School of Medicine of the University of Louisville In Partial Fulfillment of the Requirement for the Degree of Master of Science In Pharmacology and Toxicology Department of Pharmacology and Toxicology School of Medicine University of Louisville Louisville, KY August, 2016 MULTIPURPOSE TENOFOVIR DISOPROXIL FUMARATE ELECTROSPUN FIBERS FOR THE PREVENTION OF HIV-1 AND HSV-2 INFECTIONS By Kevin Tyo B.S. -

Efficacy and Safety of Imiquimod for Verruca Planae: a Systematic Review
Global Dermatology Research Article ISSN: 2056-7863 Efficacy and safety of imiquimod for verruca planae: A systematic review Xin-rui Zhang#, Bi-huan Xiao#, Rui-qun Qi*, and Xing-hua Gao* Department of Dermatology, No 1 Hospital of China Medical University, Shenyang 110001, PR China #These authors contributed equally to this work Abstract Objective: To assess the efficacy and safety of imiquimod for treating verruca planae. Methods: We searched the Pubmed, Cochrane Register of Controlled Trials, EMbase, CBM, CNKI and Wanfang databases (Chinese) to collect randomized controlled trials (RCTs). We screened the retrieved studies according to the predefined inclusion and exclusion criteria, evaluated the quality of include studies, and performed meta-analyses using the Cochrane Collaboration’s RevMan 5.1. Software. Results: Twenty-six RCTs involving 2169 patients with verruca planae were included and assessed. At the end of the 6th and ≥8th week, the effective rate of topical imiquimod was obviously higher than that of control [ RR=1.42, 95%CI (1.27, 1.60), P <0.00001; RR=1.43, 95%CI (1.22,1.67), P<0.00001]. The effective rate of imiquimod cream was higher than tretinoin cream, tazarotene gel and other antiviral drugs. [RR=1.41, 95%CI (1.25, 1.59), P <0.00001; RR=1.76, 95%CI (1.48, 2.10), P<0.00001; RR=1.71, 95%CI (1.29, 2.26), P =0.0002]. However, the effectiverate of imiquimod cream was lower than 5-ALA-PDT (RR=0.6, 95%CI (0.5, 0.71), P<0.00001). Conclusions: The limited evidence demonstrates that topical imiquimod is safe and efficient.