Containing Abasic Sites and Nicks M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(NAP) and Illustration of DNA Flexure Angles at Single Molecule Resolution Debayan Purkait†A, Debolina Bandyopadhyay†A, and Padmaja P
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.11.293639; this version posted September 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Vital Insights into Prokaryotic Genome Compaction by Nucleoid-Associated Protein (NAP) and Illustration of DNA Flexure Angles at Single Molecule Resolution Debayan Purkait†a, Debolina Bandyopadhyay†a, and Padmaja P. Mishra*a aChemical Sciences Division, Saha Institute of Nuclear Physics, India aHomi Bhaba National Institute (HBNI), India †Both the authors have contributed equally. * Corresponding author Abstract Integration Host Factor (IHF) is a heterodimeric site-specific nucleoid-associated protein (NAP) well known for its DNA bending ability. The binding is mediated through the narrow minor grooves of the consensus sequence, involving van der-Waals interaction and hydrogen bonding. Although the DNA bend state of IHF has been captured by both X-ray Crystallography and Atomic Force Microscopy (AFM), the range of flexibility and degree of heterogeneity in terms of quantitative analysis of the nucleoprotein complex has largely remained unexplored. Here we have monitored and compared the trajectories of the conformational dynamics of a dsDNA upon binding of wild-type (wt) and single-chain (sc) IHF at millisecond resolution through single-molecule FRET (smFRET). Our findings reveal that the nucleoprotein complex exists in a ‘Slacked-Dynamic’ state throughout the observation window where many of them have switched between multiple ‘Wobbling States’ in the course of attainment of packaged form. A range of DNA ‘Flexure Angles’ has been calculated that give us vital insights regarding the nucleoid organization and transcriptional regulation in prokaryotes. -

Cell Cycle Regulation by the Bacterial Nucleoid
Available online at www.sciencedirect.com ScienceDirect Cell cycle regulation by the bacterial nucleoid David William Adams, Ling Juan Wu and Jeff Errington Division site selection presents a fundamental challenge to all terminate in the terminus region (Ter; 1808). Once organisms. Bacterial cells are small and the chromosome chromosome replication and segregation are complete (nucleoid) often fills most of the cell volume. Thus, in order to the cell is ready to divide. In Bacteria this normally occurs maximise fitness and avoid damaging the genetic material, cell by binary fission and in almost all species this is initiated division must be tightly co-ordinated with chromosome by the assembly of the tubulin homologue FtsZ into a replication and segregation. To achieve this, bacteria employ a ring-like structure (‘Z-ring’) at the nascent division site number of different mechanisms to regulate division site (Figure 1) [1]. The Z-ring then functions as a dynamic selection. One such mechanism, termed nucleoid occlusion, platform for assembly of the division machinery [2,3]. Its allows the nucleoid to protect itself by acting as a template for central role in division also allows FtsZ to serve as a nucleoid occlusion factors, which prevent Z-ring assembly over regulatory hub for the majority of regulatory proteins the DNA. These factors are sequence-specific DNA-binding identified to date [2,4]. Nevertheless, the precise ultra- proteins that exploit the precise organisation of the nucleoid, structure of the Z-ring and whether or not it plays a direct allowing them to act as both spatial and temporal regulators of role in force-generation during division remains contro- bacterial cell division. -

Genetic Snapshots of the Rhizobium Species NGR234 Genome
Article Genetic snapshots of the Rhizobium species NGR234 genome VIPREY, Virginie, et al. Abstract In nitrate-poor soils, many leguminous plants form nitrogen-fixing symbioses with members of the bacterial family Rhizobiaceae. We selected Rhizobium sp. NGR234 for its exceptionally broad host range, which includes more than I 12 genera of legumes. Unlike the genome of Bradyrhizobium japonicum, which is composed of a single 8.7 Mb chromosome, that of NGR234 is partitioned into three replicons: a chromosome of about 3.5 Mb, a megaplasmid of more than 2 Mb (pNGR234b) and pNGR234a, a 536,165 bp plasmid that carries most of the genes required for symbioses with legumes. Symbiotic loci represent only a small portion of all the genes coded by rhizobial genomes, however. To rapidly characterize the two largest replicons of NGR234, the genome of strain ANU265 (a derivative strain cured of pNGR234a) was analyzed by shotgun sequencing. Reference VIPREY, Virginie, et al. Genetic snapshots of the Rhizobium species NGR234 genome. Genome Biology, 2000, vol. 1, no. 6, p. 1-17 DOI : 10.1186/gb-2000-1-6-research0014 PMID : 11178268 Available at: http://archive-ouverte.unige.ch/unige:84778 Disclaimer: layout of this document may differ from the published version. 1 / 1 http://genomebiology.com/2000/1/6/research/0014.1 Research Genetic snapshots of the Rhizobium species NGR234 genome Virginie Viprey*, André Rosenthal, William J Broughton* and Xavier Perret* Addresses: *Laboratoire de Biologie Moléculaire des Plantes Supérieures, Université de Genève, chemin de lImpératrice, 1292 Chambésy, Genève, Switzerland. Institut für Molekulare Biotechnologie, Abteilung Genomanalyze, Beutenbergstrasse, 07745 Jena, Germany. -

2.2.3 Generation of Genomic DNA Sequences from Plant Sources
Copyright Undertaking This thesis is protected by copyright, with all rights reserved. By reading and using the thesis, the reader understands and agrees to the following terms: 1. The reader will abide by the rules and legal ordinances governing copyright regarding the use of the thesis. 2. The reader will use the thesis for the purpose of research or private study only and not for distribution or further reproduction or any other purpose. 3. The reader agrees to indemnify and hold the University harmless from and against any loss, damage, cost, liability or expenses arising from copyright infringement or unauthorized usage. IMPORTANT If you have reasons to believe that any materials in this thesis are deemed not suitable to be distributed in this form, or a copyright owner having difficulty with the material being included in our database, please contact [email protected] providing details. The Library will look into your claim and consider taking remedial action upon receipt of the written requests. Pao Yue-kong Library, The Hong Kong Polytechnic University, Hung Hom, Kowloon, Hong Kong http://www.lib.polyu.edu.hk The Hong Kong Polytechnic University Department of Health Technology and Informatics Isolation and Characterisation of Nucleases from Various Biological Sources for Further Application in Nucleic Acid Analysis By CHEUNG Tsz Shan A thesis submitted in partial fulfilment of the requirements for the degree of Doctor of Philosophy March 2012 CERTIFICATE OF ORIGINALITY I hereby declare that this thesis is my own work and that, to the best of my knowledge and belief, it reproduces no materials previously published or written, nor material that has been accepted for the award of any other degree or diploma, except where due acknowledgment has been made in the text. -

Catalytic Domain of Plasmid Pad1 Relaxase Trax Defines a Group Of
Catalytic domain of plasmid pAD1 relaxase TraX defines a group of relaxases related to restriction endonucleases María Victoria Franciaa,1, Don B. Clewellb,c, Fernando de la Cruzd, and Gabriel Moncaliánd aServicio de Microbiología, Hospital Universitario Marqués de Valdecilla e Instituto de Formación e Investigación Marqués de Valdecilla, Santander 39008, Spain; bDepartment of Biologic and Materials Sciences, University of Michigan School of Dentistry, Ann Arbor, MI 48109; cDepartment of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, MI 48109; and dDepartamento de Biología Molecular e Instituto de Biomedicina y Biotecnología de Cantabria, Universidad de Cantabria–Consejo Superior de Investigaciones Científicas–Sociedad para el Desarrollo Regional de Cantabria, Santander 39011, Spain Edited by Roy Curtiss III, Arizona State University, Tempe, AZ, and approved July 9, 2013 (received for review May 30, 2013) Plasmid pAD1 is a 60-kb conjugative element commonly found in known relaxases show two characteristic sequence motifs, motif I clinical isolates of Enterococcus faecalis. The relaxase TraX and the containing the catalytic Tyr residue (which covalently attaches to primary origin of transfer oriT2 are located close to each other and the 5′ end of the cleaved DNA) and motif III with the His-triad have been shown to be essential for conjugation. The oriT2 site essential for relaxase activity (facilitating the cleavage reaction contains a large inverted repeat (where the nic site is located) by activation of the catalytic Tyr). This His-triad has been used as adjacent to a series of short direct repeats. TraX does not show a relaxase diagnostic signature (12). fi any of the typical relaxase sequence motifs but is the prototype of Plasmid pAD1 relaxase, TraX, was identi ed (9) and shown to oriT2 a unique family of relaxases (MOB ). -

Relaxation Complexes of Plasmids Colel and Cole2:Unique Site of The
Proc. Nat. Acad. Sci. USA Vol. 71, No. 10, pp. 3854-3857, October 1974 Relaxation Complexes of Plasmids ColEl and ColE2: Unique Site of the Nick in the Open Circular DNA of the Relaxed Complexes (Escherichia coli/supercoiled DNA/endonuclease/restriction enzyme/DNA replication) MICHAEL A. LOVETT, DONALD G. GUINEY, AND DONALD R. HELINSKI Department of Biology, University of California, San Diego, La Jolla, Calif. 92037 Communicated by Stanley L. Miller, June 26, 1974 ABSTRACT The product of the induced relaxation of volved in the production of a single-strand cleavage in the supercoiled DNA-protein relaxation complexes of colicino- initiation of replication and/or conjugal transfer of plasmid genic factors El (ColEl) and E2 (CoIE2) is an open circular DNA molecule with a strand-specific nick. Cleavage of the DNA. An involvement in such processes would most likely open circular DNA of each relaxed complex with the EcoRI require that the relaxation event take place at a unique site restriction endonuclease demonstrates that the single- in the DNA molecule. In this report, evidence will be pre- strand break is at a unique position. The site of the single- sented from studies using the EcoRI restriction endonuclease strand break in the relaxed ColEl complex is approximately the same distance from the EcoRI cleavage site as the that the strand-specific relaxation event takes place at a origin of ColEl DNA replication. unique site on both the ColEl and CoIE2 plasmid molecules. The location of this site, at least in the case of the ColEl Many of extrachromosomal circular DNA elements (plas- plasmid, is approximately the same distance from the single mids) of Escherichia coli, differing in molecular weight, num- EcoRI site as the origin of replication for this plasmid. -

Mung Bean Nuclease Product Information
Certificate of Analysis Mung Bean Nuclease: Part No. Size (units) Part# 9PIM431 M431A 2,000 Revised 4/18 10X Reaction Buffer (M432A): When the Mung Bean Nuclease 10X Reaction Buffer, provided with this enzyme, is diluted 1:10, it has a composition of 0.03M sodium acetate (pH 5.0), 0.05M NaCl and 1mM ZnCl2. Enzyme Storage Buffer: Mung Bean Nuclease is supplied in 10mM Tris-HCl (pH 7.5), 50mM NaCl, 0.01% Triton® X-100 and 50% glycerol. Source: Mung bean sprouts (Phaseolus aureus). Unit Definition: One unit is defined as the amount of enzyme required to produce 1µg of acid-soluble material per minute *AF9PIM431 0418M431* at 37°C. The reaction conditions are: 30mM sodium acetate (pH 5.0), 50mM NaCl, 1mM ZnCl2, 5% glycerol and 0.5mg/ml AF9PIM431 0418M431 denatured calf thymus DNA. See the unit concentration on the Product Information Label. Storage Temperature: Store at –20°C. Avoid multiple freeze-thaw cycles and exposure to frequent temperature changes. See the expiration date on the Product Information Label. Quality Control Assays Contaminant Assay Endonuclease/Nickase Activity: To confirm the absence of contaminating endonuclease/nickase activity, 1µg of lambda DNA/Hind III markers (Cat.# G1711) is incubated with Mung Bean Nuclease for 10 minutes at room temperature. The mark- ers are then separated by electrophoresis on a 1% agarose gel and stained with ethidium bromide. Markers that have been Promega Corporation incubated with ≥60 units of enzyme will remain as intact bands with a minimal amount of smearing. 2800 Woods Hollow Road Madison, WI 53711-5399 USA Telephone 608-274-4330 Toll Free 800-356-9526 Fax 608-277-2516 Internet www.promega.com PRODUCT USE LIMITATIONS, WARRANTY, DISCLAIMER Promega manufactures products for a number of intended uses. -

O-Alkyl Oligoribonucleotides As Antisense Probes (Modified RNA/Afnity Selection/Ribonudeoprotein Couiplexes/Oligonucleotide Synthesis/Biotinylation) ADOLFO M
Proc. Nail. Acad. Sci. USA Vol. 87, pp, 7747-7751, October 1990 Biochemistry 2'-O-Alkyl oligoribonucleotides as antisense probes (modified RNA/afnity selection/ribonudeoprotein couiplexes/oligonucleotide synthesis/biotinylation) ADOLFO M. IRIBARREN*, BRIAN S. SPROAT, PHILIPPE NEUNER*, INGRID SULSTON, URSULA RYDER, AND ANGUS 1. LAMOND European Molecular Biology Laboratory, Meyerhofstrasse 1, Postfach 102209, D6900 Heidelberg 1, Federal Republic of Germany Communicated by J. A. Steitz, July 3, 1990 (receivedfor review May 1, 1990) ABSTRACT 2'-O-Methyl oligoribonucleotides have re- cently been introduced as antisense probes for studying RNA processing and for affinity purification of RNA-protein com- plexes. To identify RNA analogues with improved properties for antisense analysis, 2'--alkyl oligoribonucleotides were synthesized in which the alkyl moiety was either the three- carbon linear allyl group or the five-carbon branched 3,3- dimethylallyl group. Both these analogues were found to be completely resistant to degradation by either DNA- or RNA- specific nucleases. Use of biotinylated derivatives of the probes to affinity-select ribonucleoprotein particles from crude HeLa cell nuclear extracts showed that the presence of the bulky L i3 3,3-dimethylallyl group significantly reduces affinity selection, -o'r\- .. whereas the allyl derivative binds rapidly and stably to targeted sequences and affinity-selects efficiently. The allyl derivatives also showed an increase in the level of specific binding to targeted sequences compared with 2'-0-methyl probes of iden- tical sequence. These properties indicate that the 2'-0-allyl oligoribonucleotides are particularly well suited for use as HO OR antisense probes. S H Chemically synthesized 2'-O-methyl oligoribonucleotides, as R NH described by Ohtsuka and Morisawa and coworkers (1-6) and by Sproat et al. -
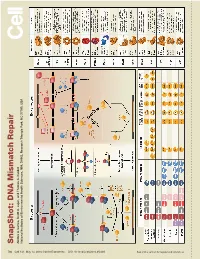
S Na P S H O T: D N a Mism a Tc H R E P a Ir
SnapShot: Repair DNA Mismatch Scott A. Lujan, and Thomas Kunkel A. Larrea, Andres Park, NC 27709, USA Triangle Health Sciences, NIH, DHHS, Research National Institutes of Environmental 730 Cell 141, May 14, 2010 ©2010 Elsevier Inc. DOI 10.1016/j.cell.2010.05.002 See online version for legend and references. SnapShot: DNA Mismatch Repair Andres A. Larrea, Scott A. Lujan, and Thomas A. Kunkel National Institutes of Environmental Health Sciences, NIH, DHHS, Research Triangle Park, NC 27709, USA Mismatch Repair in Bacteria and Eukaryotes Mismatch repair in the bacterium Escherichia coli is initiated when a homodimer of MutS binds as an asymmetric clamp to DNA containing a variety of base-base and insertion-deletion mismatches. The MutL homodimer then couples MutS recognition to the signal that distinguishes between the template and nascent DNA strands. In E. coli, the lack of adenine methylation, catalyzed by the DNA adenine methyltransferase (Dam) in newly synthesized GATC sequences, allows E. coli MutH to cleave the nascent strand. The resulting nick is used for mismatch removal involving the UvrD helicase, single-strand DNA-binding protein (SSB), and excision by single-stranded DNA exonucleases from either direction, depending upon the polarity of the nick relative to the mismatch. DNA polymerase III correctly resynthesizes DNA and ligase completes repair. In bacteria lacking Dam/MutH, as in eukaryotes, the signal for strand discrimination is uncertain but may be the DNA ends associated with replication forks. In these bacteria, MutL harbors a nick-dependent endonuclease that creates a nick that can be used for mismatch excision. Eukaryotic mismatch repair is similar, although it involves several dif- ferent MutS and MutL homologs: MutSα (MSH2/MSH6) recognizes single base-base mismatches and 1–2 base insertion/deletions; MutSβ (MSH2/MSH3) recognizes insertion/ deletion mismatches containing two or more extra bases. -

Effects of Abasic Sites and DNA Single-Strand Breaks on Prokaryotic RNA Polymerases (Apurinic/Apyrimidinic Sites/Transcription/DNA Damage) WEI ZHOU* and PAUL W
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 6601-6605, July 1993 Biochemistry Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases (apurinic/apyrimidinic sites/transcription/DNA damage) WEI ZHOU* AND PAUL W. DOETSCH*tt Departments of *Biochemistry and tRadiation Oncology and tDivision of Cancer Biology, Emory University School of Medicine, Atlanta, GA 30322 Communicated by Philip C. Hanawalt, April 19, 1993 ABSTRACT Abasic sites are thought to be the most fre- effects of such damage on both transcription initiation and quently occurring cellular DNA damage and are generated elongation processes (19, 20). We wished to determine the spontaneously or as the result of chemical or radiation damage effects of AP sites on the transcription elongation process by to DNA. In contrast to the wealth of information that exists on using a defined system that would provide direct information the effects of abasic sites on DNA polymerases, very little is regarding the ability of RNA polymerases to bypass or be known about how these lesions interact with RNA polymerases. blocked by AP sites and, if bypass was observed, to deter- An in vitro transcription system was used to determine the mine the nature of the base insertion opposite such lesions. effects of abasic sites and single-strand breaks on transcrip- For these studies, we constructed a DNA template containing tional elongation. DNA templates were constructed containing a single AP site placed at a unique location downstream from single abasic sites or nicks placed at unique locations down- either the SP6 or tac promoter and carried out in vitro stream from two different promoters and were transcribed by transcription experiments with SP6 and E. -

United States Patent (19) 11 Patent Number: 5,338,683 Paoletti (45) Date of Patent: "Aug
USOO5338683A United States Patent (19) 11 Patent Number: 5,338,683 Paoletti (45) Date of Patent: "Aug. 16, 1994 54 VACCINIAVIRUS CONTAINING DNA SEQUENCES ENCODING HERPESVIRUS FOREIGN PATENT DOCUMENTS GLY COPROTEINS 0261940 3/1988 European Pat. Off. 9001546 2/1990 PCT Int'l Appl. 75) Inventor: Enzo Paoletti, Albany, N.Y. OTHER PUBLICATIONS 73) Assignee: Health Research Incorporated, Muller et al (1977) J. Gen. Virol. 38, 135-147. Albany, N.Y. Piccini et al (1987) Meth. Enzymol 153, 545-563. Taylor et al (1988) Vaccine 6, 497-507. * Notice: The portion of the term of this patent Perkus et al (1985) Science 229, 981-984. subsequent to Jul. 29, 2003 has been Allen et al. (1987) J. Virol. 61,2454-2461. disclaimed. Piccini et al., Bioessays (Jun. 1986) vol. 5, No. 6, 248-52, at 248. 21 Appl. No.: 502,834 Elliot et al., J. Gen. Virol. (1991), 72, 1762–79, at 1763. Boyle, D. B. et al., J. Gen. Virol. (1986), 67, 1591-1600. 22 Filed: Apr. 4, 1990 Guo et al., J. of Virology, vol. 64, No. 5, pp. 2399-2406 (1990). Related U.S. Application Data Guo et al., J. of Virology, vol. 63, No. 10, pp. 4189-4198 60 Continuation-in-part of Ser. No. 394,488, Aug. 16, (1989). 1989, abandoned, and Ser. No. 90,209, Aug. 27, 1987, Primary Examiner-Elizabeth C. Weimar abandoned, which is a division of Ser. No. 622,135, Assistant Examiner-Deborah Crouch Jun. 9, 1984, Pat. No. 4,722,848, which is a continua Attorney, Agent, or Firm-Curtis, Morris & Safford tion-in-part of Ser. -

DNA Structure at the Plasmid Origin-Of-Transfer Indicates Its
www.nature.com/scientificreports OPEN DNA structure at the plasmid origin-of-transfer indicates its potential transfer range Received: 12 May 2016 Jan Zrimec1,2,3 & Aleš Lapanje1,4,5 Accepted: 10 January 2018 Horizontal gene transfer via plasmid conjugation enables antimicrobial resistance (AMR) to spread Published: xx xx xxxx among bacteria and is a major health concern. The range of potential transfer hosts of a particular conjugative plasmid is characterised by its mobility (MOB) group, which is currently determined based on the amino acid sequence of the plasmid-encoded relaxase. To facilitate prediction of plasmid MOB groups, we have developed a bioinformatic procedure based on analysis of the origin-of-transfer (oriT), a merely 230 bp long non-coding plasmid DNA region that is the enzymatic substrate for the relaxase. By computationally interpreting conformational and physicochemical properties of the oriT region, which facilitate relaxase-oriT recognition and initiation of nicking, MOB groups can be resolved with over 99% accuracy. We have shown that oriT structural properties are highly conserved and can be used to discriminate among MOB groups more efciently than the oriT nucleotide sequence. The procedure for prediction of MOB groups and potential transfer range of plasmids was implemented using published data and is available at http://dnatools.eu/MOB/plasmid.html. Antimicrobial resistance (AMR) is a pressing global issue, as it diminishes the activity of 29 antibiotics and conse- quently leads to over 25,000 deaths each year in Europe alone1,2. Te development of AMR in microbial commu- nities is facilitated by horizontal gene transfer (HGT) of conjugative elements (including plasmids and integrative elements)3 carrying antibiotic resistance genes along with virulence genes4,5.