Mutation in Archain 1, a Subunit of COPI Coatomer Complex, Causes Diluted Coat Color and Purkinje Cell Degeneration
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

Arcn1 (NM 145985) Mouse Recombinant Protein Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TP508216 Arcn1 (NM_145985) Mouse Recombinant Protein Product data: Product Type: Recombinant Proteins Description: Purified recombinant protein of Mouse archain 1 (Arcn1), with C-terminal MYC/DDK tag, expressed in HEK293T cells, 20ug Species: Mouse Expression Host: HEK293T Tag: C-MYC/DDK Predicted MW: 57.7 kDa Concentration: >50 ug/mL as determined by microplate BCA method Purity: > 80% as determined by SDS-PAGE and Coomassie blue staining Buffer: 25 mM Tris.HCl, pH 7.3, 100 mM glycine, 10% glycerol. Storage: Store at -80°C after receiving vials. Stability: Stable for 12 months from the date of receipt of the product under proper storage and handling conditions. Avoid repeated freeze-thaw cycles. RefSeq: NP_666097 Locus ID: 213827 UniProt ID: Q5XJY5 RefSeq Size: 3470 Cytogenetics: 9 A5.2 RefSeq ORF: 1536 Synonyms: 4632432M07Rik; nur17 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 Arcn1 (NM_145985) Mouse Recombinant Protein – TP508216 Summary: The coatomer is a cytosolic protein complex that binds to dilysine motifs and reversibly associates with Golgi non-clathrin-coated vesicles, which further mediate biosynthetic protein transport from the ER, via the Golgi up to the trans Golgi network. Coatomer complex is required for budding from Golgi membranes, and is essential for the retrograde Golgi-to-ER transport of dilysine-tagged proteins. -

Global Analysis of Methionine Oxidation Provides a Census Of
bioRxiv preprint doi: https://doi.org/10.1101/467290; this version posted January 10, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Global analysis of methionine oxidation provides a census of folding stabilities for the human proteome Ethan J. Walker1,2, John Q. Bettinger1, Kevin A. Welle3, Jennifer R. Hryhorenko3, Sina Ghaemmaghami1,3* 1Department of Biology, University of Rochester, NY, 14627, USA 2Department of Biochemistry, University of Rochester Medical Center, NY, 14627, USA 3University of Rochester Mass Spectrometry Resource Laboratory, NY, 14627, USA *Correspondence: [email protected] (S.G.) 1 bioRxiv preprint doi: https://doi.org/10.1101/467290; this version posted January 10, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. ABSTRACT The stability of proteins influences their tendency to aggregate, undergo degradation or become modified in cells. Despite their significance to understanding protein folding and function, quantitative analyses of thermodynamic stabilities have been mostly limited to soluble proteins in purified systems. We have used a highly multiplexed proteomics approach, based on analyses of methionine oxidation rates, to quantify stabilities of ~10,000 unique regions within ~3,000 proteins in human cell extracts. The data identify lysosomal and extracellular proteins as the most stable ontological subsets of the proteome. -
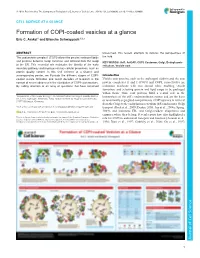
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
Supplemental Figure 2.Eps
Androgen Resveratrol 75uM 150uM 43h-10nM 43h-100nM 43h-1uM 20h-10uM 18h-25uM 20h-40uM 20h-100uM 0h 1h 3h 6h 9h 12h 15h 18h 21h 24h 48h 60h 0h 1h 2h 3h 4h 6h 8h 12h 14h 16h 18h 20h 22h 24h 48h 60h 7h-R1881 9h-R1881 18h-R1881 24h-R1881 50h-R1881 72h-R1881 18h-DHT10 50h-DHT10 24h-DHT100 24h-DHT1000 46h-noR1881 70h-noR1881 99855 ZFH4 zinc finger homeodomain 4 Hs.109314 105366 NFKB1 nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 p105 Hs.83428 112015 SEPT6 septin 6 Hs.90998 111438 SLC12A2 **solute carrier family 12 sodium/potassium/chloride transporters, member 2 Hs.110736 117460 ZIC2 Zic family member 2 odd-paired homolog, Drosophila Hs.132863 112768 KIAA0914 KIAA0914 gene product Hs.177664 111066 ESTs, Weakly similar to GNMSLL retrovirus-related reverse transcriptase homolog - mouse retrotransposon [M.musculus] Hs.301451 104992 FN1 fibronectin 1 Hs.287820 116170 RDC1 G protein-coupled receptor Hs.23016 99709 PDCD5 programmed cell death 5 Hs.166468 112624 CTBP1 C-terminal binding protein 1 Hs.239737 118259 GRB10 growth factor receptor-bound protein 10 Hs.81875 111378 PURA purine-rich element binding protein A Hs.29117 120755 **ESTs, Weakly similar to CNG1_HUMAN cGMP-gated cation channel alpha 1 CNG channel alpha 1 CNG-1 CNG1 Cyclic nucleotide gated channel alpha 1 Cyclic nucleotide gated channel, photoreceptor Hs.356525 118729 DKFZP564D0462 hypothetical protein DKFZp564D0462 Hs.44197 101077 LRBA vesicle trafficking, beach and anchor containing Hs.62354 116608 BTD biotinidase Hs.78885 107627 BCHE butyrylcholinesterase -

King's Research Portal
King’s Research Portal DOI: 10.1016/j.ajhg.2016.06.011 Document Version Peer reviewed version Link to publication record in King's Research Portal Citation for published version (APA): Izumi, K., Brett, M., Nishi, E., Drunat, S., Tan, E-S., Fujiki, K., Lebon, S., Cham, B., Masuda, K., Arakawa, M., Jacquinet, A., Yamazumi, Y., Chen, S-T., Verloes, A., Okada, Y., Katou, Y., Nakamura, T., Akiyama, T., Gressens, P., ... Shirahige, K. (2016). ARCN1 Mutations Cause a Recognizable Craniofacial Syndrome Due to COPI-Mediated Transport Defects. American Journal of Human Genetics, 99(2), 451–459. https://doi.org/10.1016/j.ajhg.2016.06.011 Citing this paper Please note that where the full-text provided on King's Research Portal is the Author Accepted Manuscript or Post-Print version this may differ from the final Published version. If citing, it is advised that you check and use the publisher's definitive version for pagination, volume/issue, and date of publication details. And where the final published version is provided on the Research Portal, if citing you are again advised to check the publisher's website for any subsequent corrections. General rights Copyright and moral rights for the publications made accessible in the Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognize and abide by the legal requirements associated with these rights. •Users may download and print one copy of any publication from the Research Portal for the purpose of private study or research. •You may not further distribute the material or use it for any profit-making activity or commercial gain •You may freely distribute the URL identifying the publication in the Research Portal Take down policy If you believe that this document breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. -

(12) United States Patent (10) Patent No.: US 7.873,482 B2 Stefanon Et Al
US007873482B2 (12) United States Patent (10) Patent No.: US 7.873,482 B2 Stefanon et al. (45) Date of Patent: Jan. 18, 2011 (54) DIAGNOSTIC SYSTEM FOR SELECTING 6,358,546 B1 3/2002 Bebiak et al. NUTRITION AND PHARMACOLOGICAL 6,493,641 B1 12/2002 Singh et al. PRODUCTS FOR ANIMALS 6,537,213 B2 3/2003 Dodds (76) Inventors: Bruno Stefanon, via Zilli, 51/A/3, Martignacco (IT) 33035: W. Jean Dodds, 938 Stanford St., Santa Monica, (Continued) CA (US) 90403 FOREIGN PATENT DOCUMENTS (*) Notice: Subject to any disclaimer, the term of this patent is extended or adjusted under 35 WO WO99-67642 A2 12/1999 U.S.C. 154(b) by 158 days. (21)21) Appl. NoNo.: 12/316,8249 (Continued) (65) Prior Publication Data Swanson, et al., “Nutritional Genomics: Implication for Companion Animals'. The American Society for Nutritional Sciences, (2003).J. US 2010/O15301.6 A1 Jun. 17, 2010 Nutr. 133:3033-3040 (18 pages). (51) Int. Cl. (Continued) G06F 9/00 (2006.01) (52) U.S. Cl. ........................................................ 702/19 Primary Examiner—Edward Raymond (58) Field of Classification Search ................... 702/19 (74) Attorney, Agent, or Firm Greenberg Traurig, LLP 702/23, 182–185 See application file for complete search history. (57) ABSTRACT (56) References Cited An analysis of the profile of a non-human animal comprises: U.S. PATENT DOCUMENTS a) providing a genotypic database to the species of the non 3,995,019 A 1 1/1976 Jerome human animal Subject or a selected group of the species; b) 5,691,157 A 1 1/1997 Gong et al. -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942 -

Supplementary Material Computational Prediction of SARS
Supplementary_Material Computational prediction of SARS-CoV-2 encoded miRNAs and their putative host targets Sheet_1 List of potential stem-loop structures in SARS-CoV-2 genome as predicted by VMir. Rank Name Start Apex Size Score Window Count (Absolute) Direct Orientation 1 MD13 2801 2864 125 243.8 61 2 MD62 11234 11286 101 211.4 49 4 MD136 27666 27721 104 205.6 119 5 MD108 21131 21184 110 204.7 210 9 MD132 26743 26801 119 188.9 252 19 MD56 9797 9858 128 179.1 59 26 MD139 28196 28233 72 170.4 133 28 MD16 2934 2974 76 169.9 71 43 MD103 20002 20042 80 159.3 403 46 MD6 1489 1531 86 156.7 171 51 MD17 2981 3047 131 152.8 38 87 MD4 651 692 75 140.3 46 95 MD7 1810 1872 121 137.4 58 116 MD140 28217 28252 72 133.8 62 122 MD55 9712 9758 96 132.5 49 135 MD70 13171 13219 93 130.2 131 164 MD95 18782 18820 79 124.7 184 173 MD121 24086 24135 99 123.1 45 176 MD96 19046 19086 75 123.1 179 196 MD19 3197 3236 76 120.4 49 200 MD86 17048 17083 73 119.8 428 223 MD75 14534 14600 137 117 51 228 MD50 8824 8870 94 115.8 79 234 MD129 25598 25642 89 115.6 354 Reverse Orientation 6 MR61 19088 19132 88 197.8 271 10 MR72 23563 23636 148 188.8 286 11 MR11 3775 3844 136 185.1 116 12 MR94 29532 29582 94 184.6 271 15 MR43 14973 15028 109 183.9 226 27 MR14 4160 4206 89 170 241 34 MR35 11734 11792 111 164.2 37 52 MR5 1603 1652 89 152.7 118 53 MR57 18089 18132 101 152.7 139 94 MR8 2804 2864 122 137.4 38 107 MR58 18474 18508 72 134.9 237 117 MR16 4506 4540 72 133.8 311 120 MR34 10010 10048 82 132.7 245 133 MR7 2534 2578 90 130.4 75 146 MR79 24766 24808 75 127.9 59 150 MR65 21528 21576 99 127.4 83 180 MR60 19016 19049 70 122.5 72 187 MR51 16450 16482 75 121 363 190 MR80 25687 25734 96 120.6 75 198 MR64 21507 21544 70 120.3 35 206 MR41 14500 14542 84 119.2 94 218 MR84 26840 26894 108 117.6 94 Sheet_2 List of stable stem-loop structures based on MFE. -

Target Gene Gene Description Validation Diana Miranda
Supplemental Table S1. Mmu-miR-183-5p in silico predicted targets. TARGET GENE GENE DESCRIPTION VALIDATION DIANA MIRANDA MIRBRIDGE PICTAR PITA RNA22 TARGETSCAN TOTAL_HIT AP3M1 adaptor-related protein complex 3, mu 1 subunit V V V V V V V 7 BTG1 B-cell translocation gene 1, anti-proliferative V V V V V V V 7 CLCN3 chloride channel, voltage-sensitive 3 V V V V V V V 7 CTDSPL CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like V V V V V V V 7 DUSP10 dual specificity phosphatase 10 V V V V V V V 7 MAP3K4 mitogen-activated protein kinase kinase kinase 4 V V V V V V V 7 PDCD4 programmed cell death 4 (neoplastic transformation inhibitor) V V V V V V V 7 PPP2R5C protein phosphatase 2, regulatory subunit B', gamma V V V V V V V 7 PTPN4 protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) V V V V V V V 7 EZR ezrin V V V V V V 6 FOXO1 forkhead box O1 V V V V V V 6 ANKRD13C ankyrin repeat domain 13C V V V V V V 6 ARHGAP6 Rho GTPase activating protein 6 V V V V V V 6 BACH2 BTB and CNC homology 1, basic leucine zipper transcription factor 2 V V V V V V 6 BNIP3L BCL2/adenovirus E1B 19kDa interacting protein 3-like V V V V V V 6 BRMS1L breast cancer metastasis-suppressor 1-like V V V V V V 6 CDK5R1 cyclin-dependent kinase 5, regulatory subunit 1 (p35) V V V V V V 6 CTDSP1 CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 V V V V V V 6 DCX doublecortin V V V V V V 6 ENAH enabled homolog (Drosophila) V V V V V V 6 EPHA4 EPH receptor A4 V V V V V V 6 FOXP1 forkhead box P1 V