A SAM Oligomerization Domain Shapes the Genomic Binding Landscape of the LEAFY Transcription Factor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Arabidopsis LEAFY COTYLEDON2 Induces Maturation Traits and Auxin Activity: Implications for Somatic Embryogenesis
Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis Sandra L. Stone*, Siobhan A. Braybrook*†, Stephanie L. Paula*‡, Linda W. Kwong*, Jonathan Meuser*§, Julie Pelletier*, Tzung-Fu Hsieh¶, Robert L. Fischer¶, Robert B. Goldbergʈ**, and John J. Harada*†** *Section of Plant Biology, College of Biological Sciences, and †Graduate Program in Plant Biology, University of California, Davis, CA 95616; ¶Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720; and ʈDepartment of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, CA 90024 Contributed by Robert B. Goldberg, December 30, 2007 (sent for review December 16, 2007) LEAFY COTYLEDON2 (LEC2) is a central regulator of embryogenesis INSENSITIVE3 (ABI3) and another LEC protein, FUSCA3 sufficient to induce somatic cells to form embryos when expressed (FUS3), also play critical roles in embryogenesis (11, 12). ectopically. Here, we analyze the cellular processes induced by To gain insight into the mechanisms by which cells change LEC2, a B3 domain transcription factor, that may underlie its ability their fate and become embryogenic, we analyzed postembryonic to promote somatic embryogenesis. We show auxin-responsive 35S:LEC2 plants. We showed previously LEC2 is expressed genes are induced after LEC2 activation in seedlings. Genes en- primarily during seed development and ectopic postembryonic coding enzymes involved in auxin biosynthesis, YUC2 and YUC4, expression of LEC2 induces vegetative cells to undergo somatic are activated within 1 h after induction of LEC2 activity, and YUC4 embryogenesis (8). We hypothesized LEC2 establishes a cellular appears to be a direct transcriptional target of LEC2. We also show environment that promotes embryo development. -

Upregulation of a KN1 Homolog by Transposon Insertion Promotes Leafy Head Development in Lettuce
Upregulation of a KN1 homolog by transposon insertion promotes leafy head development in lettuce Changchun Yua,1, Chenghuan Yana,1, Yuling Liua, Yali Liua, Yue Jiaa, Dean Lavelleb, Guanghui Ana, Weiyi Zhanga, Lei Zhanga, Rongkui Hanb, Robert M. Larkina, Jiongjiong Chena, Richard W. Michelmoreb, and Hanhui Kuanga,2 aKey Laboratory of Horticultural Plant Biology, Ministry of Education, Key Laboratory of Horticultural Crop Biology and Genetic improvement (Central Region), Ministry of Agriculture (MOA), College of Horticulture and Forestry Sciences, Huazhong Agricultural University, 430070 Wuhan, People’s Republic of China; and bGenome Center and Department of Plant Sciences, University of California, Davis, CA 95616 Edited by Martin F. Yanofsky, University of California San Diego, La Jolla, CA, and approved November 4, 2020 (received for review September 19, 2020) Leafy head is a unique type of plant architecture found in some axes. Heading in vegetable crops is considered to be a develop- vegetable crops, with leaves bending inward to form a compact head. mental disorder of leaf dorsoventrality (i.e., incongruous cell di- The genetic and molecular mechanisms underlying leafy head in vision/expansion along the adaxial–abaxial axis). Genes involved in vegetables remain poorly understood. We genetically fine-mapped leaf dorsoventrality include AS1, AS2, miR390, TAS3, KAN, and cloned a major quantitative trait locus controlling heading in YABBY, ARF3, ARF4, PHB,andREV (6–12). Genes associated lettuce. The candidate gene (LsKN1)isahomologofknotted 1 with dorsoventrality are directly or indirectly regulated by key (KN1)fromZea mays. Complementation and CRISPR/Cas9 knockout developmental regulators of leaves, such as members of the experiments confirmed the role of LsKN1 in heading. -

LEAFY, a Homeotic Gene That Regulates Lnflorescence Development in Arabidopsis
The Plant Cell, Vol. 3, 771 -781, August 1991 o 1991 American Society of Plant Physiologists LEAFY, a Homeotic Gene That Regulates lnflorescence Development in Arabidopsis Elizabeth A. Schultz and George W. Haughn’ Biology Department, University of Saskatchewan, Saskatoon, Saskatchewan, S7N OWO, Canada Variation in plant shoot structure may be described as occurring through changes within a basic unit, the metamer. Using this terminology, the apical meristem of Arabidopsis produces three metameric types sequentially: type 1, rosette; type 2, coflorescence-bearing with bract; and type 3, flower-bearing without bract. We describe a mutant of Arabidopsis, Leafy, homozygous for a recessive allele of a nuclear gene LEAFY (LFY), that has an inflorescence composed only of type 2-like metamers. These data suggest that the LFY gene is required for the development of type 3 metamers and that the transition from type 2 to type 3 metamers is a developmental step distinct from that between vegetative and reproductive growth (type 1 to type 2 metamers). Results from double mutant analysis, showing that /fy-7 is epistatic to the floral organ homeotic gene ap2-6, are consistent with the hypothesis that a functional LFY gene is necessary for the expression of downstream genes controlling floral organ identity. INTRODUCTION Angiosperm shoots can be described as a series of re- pattern of development of the main inflorescence, giving peating units (metamers) that are formed sequentially by rise itself to lateral coflorescenceshoots and flowers. Thus, the apical meristem. In the most basic form, a metamer the inflorescence of Arabidopsis should form an infinite consists of a node with the associated leaflike organ, the series of coflorescences if unlimited growth were possible. -

Interactions Among APETALA1, LEAFY, and TERMINAL FLOWER1 Specify Meristem Fate
The Plant Cell, Vol. 11, 1007–1018, June 1999, www.plantcell.org © 1999 American Society of Plant Physiologists RESEARCH ARTICLE Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 Specify Meristem Fate Sarah J. Liljegren, Cindy Gustafson-Brown, Anusak Pinyopich, Gary S. Ditta, and Martin F. Yanofsky1 Department of Biology and Center for Molecular Genetics, University of California at San Diego, La Jolla, California 92093 Upon floral induction, the primary shoot meristem of an Arabidopsis plant begins to produce flower meristems rather than leaf primordia on its flanks. Assignment of floral fate to lateral meristems is primarily due to the cooperative activ- ity of the flower meristem identity genes LEAFY (LFY), APETALA1 (AP1), and CAULIFLOWER. We present evidence here that AP1 expression in lateral meristems is activated by at least two independent pathways, one of which is regulated by LFY. In lfy mutants, the onset of AP1 expression is delayed, indicating that LFY is formally a positive regulator of AP1. We have found that AP1, in turn, can positively regulate LFY, because LFY is expressed prematurely in the con- verted floral meristems of plants constitutively expressing AP1. Shoot meristems maintain an identity distinct from that of flower meristems, in part through the action of genes such as TERMINAL FLOWER1 (TFL1), which bars AP1 and LFY expression from the inflorescence shoot meristem. We show here that this negative regulation can be mutual because TFL1 expression is downregulated in plants constitutively expressing AP1. Therefore, the normally sharp phase transi- tion between the production of leaves with associated shoots and formation of the flowers, which occurs upon floral in- duction, is promoted by positive feedback interactions between LFY and AP1, together with negative interactions of these two genes with TFL1. -

LEAFY Maintains Apical Stem Cell Activity During Shoot Development In
RESEARCH ARTICLE LEAFY maintains apical stem cell activity during shoot development in the fern Ceratopteris richardii Andrew RG Plackett1†‡, Stephanie J Conway2†§, Kristen D Hewett Hazelton2, Ester H Rabbinowitsch1, Jane A Langdale1*, Vero´ nica S Di Stilio2* 1Department of Plant Sciences, University of Oxford, Oxford, United Kingdom; 2Department of Biology, University of Washington, Seattle, United States Abstract During land plant evolution, determinate spore-bearing axes (retained in extant bryophytes such as mosses) were progressively transformed into indeterminate branching shoots with specialized reproductive axes that form flowers. The LEAFY transcription factor, which is required for the first zygotic cell division in mosses and primarily for floral meristem identity in flowering plants, may have facilitated developmental innovations during these transitions. Mapping *For correspondence: the LEAFY evolutionary trajectory has been challenging, however, because there is no functional [email protected] overlap between mosses and flowering plants, and no functional data from intervening lineages. (JAL); [email protected] (VSD) Here, we report a transgenic analysis in the fern Ceratopteris richardii that reveals a role for LEAFY in maintaining cell divisions in the apical stem cells of both haploid and diploid phases of the † These authors contributed lifecycle. These results support an evolutionary trajectory in which an ancestral LEAFY module that equally to this work promotes cell proliferation was progressively co-opted, -

Diversity and Evolution of the Megaphyll in Euphyllophytes
G Model PALEVO-665; No. of Pages 16 ARTICLE IN PRESS C. R. Palevol xxx (2012) xxx–xxx Contents lists available at SciVerse ScienceDirect Comptes Rendus Palevol w ww.sciencedirect.com General palaeontology, systematics and evolution (Palaeobotany) Diversity and evolution of the megaphyll in Euphyllophytes: Phylogenetic hypotheses and the problem of foliar organ definition Diversité et évolution de la mégaphylle chez les Euphyllophytes : hypothèses phylogénétiques et le problème de la définition de l’organe foliaire ∗ Adèle Corvez , Véronique Barriel , Jean-Yves Dubuisson UMR 7207 CNRS-MNHN-UPMC, centre de recherches en paléobiodiversité et paléoenvironnements, 57, rue Cuvier, CP 48, 75005 Paris, France a r t i c l e i n f o a b s t r a c t Article history: Recent paleobotanical studies suggest that megaphylls evolved several times in land plant st Received 1 February 2012 evolution, implying that behind the single word “megaphyll” are hidden very differ- Accepted after revision 23 May 2012 ent notions and concepts. We therefore review current knowledge about diverse foliar Available online xxx organs and related characters observed in fossil and living plants, using one phylogenetic hypothesis to infer their origins and evolution. Four foliar organs and one lateral axis are Presented by Philippe Taquet described in detail and differ by the different combination of four main characters: lateral organ symmetry, abdaxity, planation and webbing. Phylogenetic analyses show that the Keywords: “true” megaphyll appeared at least twice in Euphyllophytes, and that the history of the Euphyllophytes Megaphyll four main characters is different in each case. The current definition of the megaphyll is questioned; we propose a clear and accurate terminology in order to remove ambiguities Bilateral symmetry Abdaxity of the current vocabulary. -

LEAFY COTYLEDON2 Encodes a B3 Domain Transcription Factor That Induces Embryo Development
LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development Sandra L. Stone*†, Linda W. Kwong*†, Kelly Matsudaira Yee*, Julie Pelletier*, Loı¨c Lepiniec‡, Robert L. Fischer§, Robert B. Goldberg¶, and John J. Harada*ʈ *Section of Plant Biology, Division of Biological Sciences, University of California, One Shields Avenue, Davis, CA 95616; ‡Seed Biology Laboratory, Laboratoire de Biologie des Semences, Institut National de la Recherche Agronomique, Route de St-Cyr, 78026 Versailles, France; §Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720; and ¶Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, CA 90024-1606 Contributed by Robert B. Goldberg, August 6, 2001 The Arabidopsis LEAFY COTYLEDON2(LEC2) gene is a central Early in embryogenesis, LEC genes are required to specify embryonic regulator that serves critical roles both early and late suspensor cell fate and cotyledon identity (12–16). Late in during embryo development. LEC2 is required for the maintenance embryogenesis, LEC genes are needed during the maturation of suspensor morphology, specification of cotyledon identity, phase for the acquisition of desiccation tolerance and the progression through the maturation phase, and suppression of expression of many maturation-specific genes (13–17). Consis- premature germination. We cloned the LEC2 gene on the basis of tent with the finding that conditions that promote maturation its chromosomal position and showed that the predicted polypep- suppress germination (6), lec mutant embryos prematurely ac- tide contains a B3 domain, a DNA-binding motif unique to plants tivate the postgermination program (13, 15, 16, 18). Thus, LEC that is characteristic of several transcription factors. -

Flowers Into Shoots: Photo and Hormonal Control of a Meristem Identity Switch in Arabidopsis (Gibberellins͞phytochrome͞floral Reversion)
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 13831–13836, November 1996 Developmental Biology Flowers into shoots: Photo and hormonal control of a meristem identity switch in Arabidopsis (gibberellinsyphytochromeyfloral reversion) JACK K. OKAMURO*, BART G. W. DEN BOER†,CYNTHIA LOTYS-PRASS,WAYNE SZETO, AND K. DIANE JOFUKU Department of Biology, University of California, Santa Cruz, CA 95064 Communicated by Bernard O. Phinney, University of California, Los Angeles, CA, September 10, 1996 (received for review February 1, 1996) ABSTRACT Little is known about the signals that govern MOUS (AG) (3, 15, 17, 21–23). The LFY gene encodes a novel the network of meristem and organ identity genes that control polypeptide that is reported to have DNA binding activity in flower development. In Arabidopsis, we can induce a hetero- vitro (20). AG encodes a protein that belongs to the evolu- chronic switch from flower to shoot development, a process tionarily conserved MADS domain family of eukaryotic tran- known as floral meristem reversion, by manipulating photo- scription factors (24, 25). However, little is known about how period in the floral homeotic mutant agamous and in plants these proteins govern meristem identity at the cellular and heterozygous for the meristem identity gene leafy. The trans- molecular levels. Previous studies suggest that the mainte- formation from flower to shoot meristem is suppressed by hy1, nance of flower meristem identity in lfy and in ag mutants is a mutation blocking phytochrome activity, by spindly, a mu- controlled by photoperiod and by one class of plant growth tation that activates basal gibberellin signal transduction in regulators, the gibberellins (12, 15, 21). -
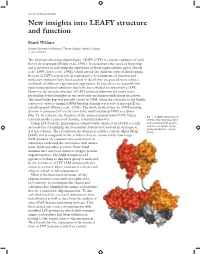
New Insights Into LEAFY Structure and Function
doi:10.1093/jxb/erx092 New insights into LEAFY structure and function Frank Wellmer Smurfit Institute of Genetics, Trinity College Dublin, Ireland [email protected] The plant-specific transcription factor LEAFY (LFY) is a master regulator of early flower development (Weigel et al., 1992). It orchestrates the onset of flowering and is involved in activating the expression of floral organ identity genes (Busch et al., 1999; Parcy et al., 1998), which specify the different types of floral organs. Because of LFY’s central role in reproductive development, its function and molecular evolution have been studied in detail over the past 25 years using a multitude of different experimental approaches. In fact, there are arguably few plant transcriptional regulators that have been studied as extensively as LFY. However, the protein structure of LFY remained unknown for many years, precluding detailed insights in the molecular mechanism underlying its activity. This knowledge gap was partially closed in 2008, when the structure of the highly conserved carboxy-terminal DNA binding domain was resolved through X-ray crystallography (Hamès et al., 2008). This work showed that the DNA binding domain is composed of a helix-turn-helix motif and binds DNA as a dimer (Fig. 1). In contrast, the function of the amino-terminal half of LFY, which Fig. 1. A LEAFY dimer bound contains another conserved domain, remained unknown. to DNA. The N-terminal SAM Using LFY from the gymnosperm Gingko biloba, Sayou et al. (2016) recently and C-terminal DNA binding succeeded in crystallizing the N-terminal domain and resolved its structure at domains are shown. -

Genomic Identification of Direct Target Genes of LEAFY
Genomic identification of direct target genes of LEAFY Dilusha A. William*, Yanhui Su*, Michael R. Smith*, Meina Lu*, Don A. Baldwin†, and Doris Wagner*‡ *Department of Biology and †Penn Microarray Facility, University of Pennsylvania, 415 South University Avenue, Philadelphia, PA 19104 Communicated by Anthony R. Cashmore, University of Pennsylvania, Philadelphia, PA, November 25, 2003 (received for review September 5, 2003) The switch from vegetative to reproductive development in plants AP1 acts as a floral homeotic gene in addition to its role as a necessitates a switch in the developmental program of the descen- meristem identity regulator (17, 18). The latter function does not dents of the stem cells in the shoot apical meristem. Genetic and depend on LFY, however, because lfy null mutants show strong AP1 molecular investigations have demonstrated that the plant-specific expression in flowers from stage three onward (11, 13, 16, 19). The transcription factor and meristem identity regulator LEAFY (LFY) LFY-independent induction of AP1 expression makes it difficult to controls this developmental transition by inducing expression of a detect LFY-dependent induction of AP1 at the floral transition by second transcription factor, APETALA1, and by regulating the expres- using quantitative methods in entire inflorescences (13). To identify sion of additional, as yet unknown, genes. Here we show that the the additional unknown LFY targets that act at the transition to additional LFY targets include the APETALA1-related factor, CAULI- reproductive development together with AP1, it is, therefore, FLOWER, as well as three transcription factors and two putative signal important to first identify a developmental stage at which quanti- transduction pathway components. -

A Role for Late Meristem Identity2 in the Reproductive Development of Arabidopsis
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations Summer 2011 A Role for Late Meristem Identity2 in the Reproductive Development of Arabidopsis Jennifer J. Pastore University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Biology Commons, Developmental Biology Commons, and the Plant Biology Commons Recommended Citation Pastore, Jennifer J., "A Role for Late Meristem Identity2 in the Reproductive Development of Arabidopsis" (2011). Publicly Accessible Penn Dissertations. 981. https://repository.upenn.edu/edissertations/981 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/981 For more information, please contact [email protected]. A Role for Late Meristem Identity2 in the Reproductive Development of Arabidopsis Abstract The switch from producing vegetative structures--branches and leaves--to producing reproductive structures--flowers--is a crucial developmental transition that significantly affects the reproductive success of flowering plants. In Arabidopsis thaliana , this transition is in large part controlled by the meristem identity regulator LEAFY (LFY) and the LFY direct target APETALA1 (AP1 ). The molecular mechanisms by which LFY orchestrates a precise and robust switch to flower formation is not well understood. Here we show that the R2R3 MYB transcription factor and direct LFY target LATE MERISTEM IDENTITY2 ( LMI2 ) plays a role in the meristem identity transition. Like LFY, LMI2 directly activates AP1 ; moreover LMI2 and LFY physically interact. LFY, LMI2 and AP1 are connected in a feed-forward and positive feedback loop network. We propose that these intricate regulatory interactions direct not only the precision of this critical developmental transition, but also contribute to its robustness and irreversibility. -

Genetic Containment in Vegetatively Propagated Forest Trees: CRISPR
Plant Biotechnology Journal (2021), pp. 1–13 doi: 10.1111/pbi.13588 Genetic containment in vegetatively propagated forest trees: CRISPR disruption of LEAFY function in Eucalyptus gives sterile indeterminate inflorescences and normal juvenile development Estefania Elorriaga1,a, Amy L. Klocko2, Cathleen Ma1, Marc du Plessis3, Xinmin An4, Alexander A. Myburg5 and Steven H. Strauss1,* 1Department of Forest Ecosystems and Society, Oregon State University, Corvallis, OR, USA 2Department of Biology, University of Colorado Colorado Springs, Colorado Springs, CO, USA 3Department of Zoology and Entomology, University of Pretoria, Pretoria, South Africa 4Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, National Engineering Laboratory for Tree Breeding, College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China 5Department of Biochemistry, Genetics and Microbiology, Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa Received 16 December 2020; Summary revised 27 February 2021; Eucalyptus is among the most widely planted taxa of forest trees worldwide. However, its spread accepted 14 March 2021. as an exotic or genetically engineered form can create ecological and social problems. To + *Correspondence (Tel 1 541-737-6578; fax mitigate gene flow via pollen and seeds, we mutated the Eucalyptus orthologue of LEAFY (LFY) +1 541-737-1393; email: by transforming a Eucalyptus grandis 9 urophylla hybrid and two Flowering Locus T (FT) [email protected]) overexpressing (and flowering) lines with CRISPR Cas9 targeting its LFY orthologue, ELFY.We aPresent address: Department of Molecular and Structural Biochemistry, North Carolina achieved high rates of elfy biallelic knockouts, often approaching 100% of transgene insertion State University, Raleigh, NC, USA events.