Gene Expression Analysis of Drosophilaa Manf Mutants Reveals
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

A Novel De Novo 20Q13.32&Ndash;Q13.33
Journal of Human Genetics (2015) 60, 313–317 & 2015 The Japan Society of Human Genetics All rights reserved 1434-5161/15 www.nature.com/jhg ORIGINAL ARTICLE Anovelde novo 20q13.32–q13.33 deletion in a 2-year-old child with poor growth, feeding difficulties and low bone mass Meena Balasubramanian1, Edward Atack2, Kath Smith2 and Michael James Parker1 Interstitial deletions of the long arm of chromosome 20 are rarely reported in the literature. We report a 2-year-old child with a 2.6 Mb deletion of 20q13.32–q13.33, detected by microarray-based comparative genomic hybridization, who presented with poor growth, feeding difficulties, abnormal subcutaneous fat distribution with the lack of adipose tissue on clinical examination, facial dysmorphism and low bone mass. This report adds to rare publications describing constitutional aberrations of chromosome 20q, and adds further evidence to the fact that deletion of the GNAS complex may not always be associated with an Albright’s hereditary osteodystrophy phenotype as described previously. Journal of Human Genetics (2015) 60, 313–317; doi:10.1038/jhg.2015.22; published online 12 March 2015 INTRODUCTION resuscitation immediately after birth and Apgar scores were 9 and 9 at 1 and Reports of isolated subtelomeric deletions of the long arm of 10 min, respectively, of age. Birth parameters were: weight ~ 1.56 kg (0.4th–2nd chromosome 20 are rare, but a few cases have been reported in the centile), length ~ 40 cm (o0.4th centile) and head circumference ~ 28.2 cm o fi literature over the past 30 years.1–13 Traylor et al.12 provided an ( 0.4th centile). -

ATP5E (A-11): Sc-393695
SANTA CRUZ BIOTECHNOLOGY, INC. ATP5E (A-11): sc-393695 BACKGROUND APPLICATIONS Mitochondrial ATP synthases (ATPases) transduce the energy contained in ATP5E (A-11) is recommended for detection of ATP5E of human origin membrane electrochemical proton gradients into the energy required for syn- by Western Blotting (starting dilution 1:100, dilution range 1:100-1:1000), thesis of high-energy phosphate bonds. ATPases contain two linked complexes: immunoprecipitation [1-2 µg per 100-500 µg of total protein (1 ml of cell F1, the hydrophilic catalytic core; and F0, the membrane-embedded protein lysate)], immunofluorescence (starting dilution 1:50, dilution range 1:50- channel. F1 consists of three a chains and three b chains, which are weakly 1:500) and solid phase ELISA (starting dilution 1:30, dilution range 1:30- homologous, as well as one g chain, one d chain and one e chain. F0 consists 1:3000). of three subunits: a, b and c. The chain of F contains 50 amino acids and e 1 Suitable for use as control antibody for ATP5E siRNA (h): sc-60229, ATP5E is the smallest of the 5 ATPase F chains. Mitochondrial ATPase chain 1 e shRNA Plasmid (h): sc-60229-SH and ATP5E shRNA (h) Lentiviral Particles: (ATP5E) localizes to the mitochondria and catalyzes ATP synthesis. sc-60229-V. REFERENCES Molecular Weight of ATP5E: 7 kDa. 1. Walker, J.E., et al. 1985. Primary structure and subunit stoichiometry of Positive Controls: HeLa whole cell lysate: sc-2200 or SW-13 cell lysate: F1-ATPase from bovine mitochondria. J. Mol. Biol. 184: 677-701. -

The DNA Sequence and Comparative Analysis of Human Chromosome 20
articles The DNA sequence and comparative analysis of human chromosome 20 P. Deloukas, L. H. Matthews, J. Ashurst, J. Burton, J. G. R. Gilbert, M. Jones, G. Stavrides, J. P. Almeida, A. K. Babbage, C. L. Bagguley, J. Bailey, K. F. Barlow, K. N. Bates, L. M. Beard, D. M. Beare, O. P. Beasley, C. P. Bird, S. E. Blakey, A. M. Bridgeman, A. J. Brown, D. Buck, W. Burrill, A. P. Butler, C. Carder, N. P. Carter, J. C. Chapman, M. Clamp, G. Clark, L. N. Clark, S. Y. Clark, C. M. Clee, S. Clegg, V. E. Cobley, R. E. Collier, R. Connor, N. R. Corby, A. Coulson, G. J. Coville, R. Deadman, P. Dhami, M. Dunn, A. G. Ellington, J. A. Frankland, A. Fraser, L. French, P. Garner, D. V. Grafham, C. Grif®ths, M. N. D. Grif®ths, R. Gwilliam, R. E. Hall, S. Hammond, J. L. Harley, P. D. Heath, S. Ho, J. L. Holden, P. J. Howden, E. Huckle, A. R. Hunt, S. E. Hunt, K. Jekosch, C. M. Johnson, D. Johnson, M. P. Kay, A. M. Kimberley, A. King, A. Knights, G. K. Laird, S. Lawlor, M. H. Lehvaslaiho, M. Leversha, C. Lloyd, D. M. Lloyd, J. D. Lovell, V. L. Marsh, S. L. Martin, L. J. McConnachie, K. McLay, A. A. McMurray, S. Milne, D. Mistry, M. J. F. Moore, J. C. Mullikin, T. Nickerson, K. Oliver, A. Parker, R. Patel, T. A. V. Pearce, A. I. Peck, B. J. C. T. Phillimore, S. R. Prathalingam, R. W. Plumb, H. Ramsay, C. M. -

Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces Cerevisiae
life Review Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae Leticia V. R. Franco 1,2,* , Luca Bremner 1 and Mario H. Barros 2 1 Department of Biological Sciences, Columbia University, New York, NY 10027, USA; [email protected] 2 Department of Microbiology,Institute of Biomedical Sciences, Universidade de Sao Paulo, Sao Paulo 05508-900, Brazil; [email protected] * Correspondence: [email protected] Received: 27 October 2020; Accepted: 19 November 2020; Published: 23 November 2020 Abstract: The ease with which the unicellular yeast Saccharomyces cerevisiae can be manipulated genetically and biochemically has established this organism as a good model for the study of human mitochondrial diseases. The combined use of biochemical and molecular genetic tools has been instrumental in elucidating the functions of numerous yeast nuclear gene products with human homologs that affect a large number of metabolic and biological processes, including those housed in mitochondria. These include structural and catalytic subunits of enzymes and protein factors that impinge on the biogenesis of the respiratory chain. This article will review what is currently known about the genetics and clinical phenotypes of mitochondrial diseases of the respiratory chain and ATP synthase, with special emphasis on the contribution of information gained from pet mutants with mutations in nuclear genes that impair mitochondrial respiration. Our intent is to provide the yeast mitochondrial specialist with basic knowledge of human mitochondrial pathologies and the human specialist with information on how genes that directly and indirectly affect respiration were identified and characterized in yeast. Keywords: mitochondrial diseases; respiratory chain; yeast; Saccharomyces cerevisiae; pet mutants 1. -

Motor Neurone Disease Variant at 20Q13 J Med Genet: First Published As on 1 April 2004
315 LETTER TO JMG A novel locus for late onset amyotrophic lateral sclerosis/ motor neurone disease variant at 20q13 J Med Genet: first published as on 1 April 2004. Downloaded from A L Nishimura, M Mitne-Neto, H C A Silva, J R M Oliveira, M Vainzof, M Zatz ............................................................................................................................... J Med Genet 2004;41:315–320. doi: 10.1136/jmg.2003.013029 otor neurone disease includes a heterogeneous group Key points of disorders with motor neurone involvement, such as Mamyotrophic lateral sclerosis, progressive muscular N Motor neurone disease includes a heterogeneous atrophy, progressive bulbar palsy, and primary lateral group of disorders with motor neurone involvement, sclerosis. Amyotrophic lateral sclerosis is the most common such as amyotrophic lateral sclerosis, progressive adult onset form of motor neurone disease and involves the muscular atrophy, progressive bulbar palsy, and lower and upper motor neurones. It is characterised by primary lateral sclerosis. progressive muscle weakness and atrophy, with fascicula- N Amyotrophic lateral sclerosis is the most common adult tions associated with hyperreflexia and spasticity. One of the proposed mechanisms for amyotrophic lateral sclerosis is onset form of motor neurone disease and involves degeneration of the motor neurone because of abnormal lower and upper motor neurones. It is characterised by levels of toxic products that accumulate in the cell. Death progressive muscle weakness and atrophy, with fasci- usually occurs by respiratory failure about 2–3 years after the culations associated with hyperreflexia and spasticity. first symptoms.1 About 10% of cases are familial amyotrophic N One proposed mechanism for amyotrophic lateral lateral sclerosis, and several loci have been associated with sclerosis is degeneration of the motor neurone because this disease. -
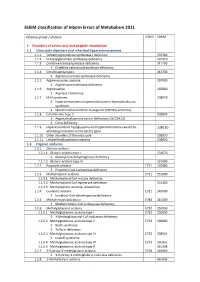
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions
International Journal of Molecular Sciences Review Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions Margherita Protasoni 1 and Massimo Zeviani 1,2,* 1 Mitochondrial Biology Unit, The MRC and University of Cambridge, Cambridge CB2 0XY, UK; [email protected] 2 Department of Neurosciences, University of Padova, 35128 Padova, Italy * Correspondence: [email protected] Abstract: Mitochondria are ubiquitous intracellular organelles found in almost all eukaryotes and involved in various aspects of cellular life, with a primary role in energy production. The interest in this organelle has grown stronger with the discovery of their link to various pathologies, including cancer, aging and neurodegenerative diseases. Indeed, dysfunctional mitochondria cannot provide the required energy to tissues with a high-energy demand, such as heart, brain and muscles, leading to a large spectrum of clinical phenotypes. Mitochondrial defects are at the origin of a group of clinically heterogeneous pathologies, called mitochondrial diseases, with an incidence of 1 in 5000 live births. Primary mitochondrial diseases are associated with genetic mutations both in nuclear and mitochondrial DNA (mtDNA), affecting genes involved in every aspect of the organelle function. As a consequence, it is difficult to find a common cause for mitochondrial diseases and, subsequently, to offer a precise clinical definition of the pathology. Moreover, the complexity of this condition makes it challenging to identify possible therapies or drug targets. Keywords: ATP production; biogenesis of the respiratory chain; mitochondrial disease; mi-tochondrial electrochemical gradient; mitochondrial potential; mitochondrial proton pumping; mitochondrial respiratory chain; oxidative phosphorylation; respiratory complex; respiratory supercomplex Citation: Protasoni, M.; Zeviani, M. -

Tissue-Specific Disallowance of Housekeeping Genes
Downloaded from genome.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Tissue-specific disallowance of housekeeping genes: the other face of cell differentiation Lieven Thorrez1,2,4, Ilaria Laudadio3, Katrijn Van Deun4, Roel Quintens1,4, Nico Hendrickx1,4, Mikaela Granvik1,4, Katleen Lemaire1,4, Anica Schraenen1,4, Leentje Van Lommel1,4, Stefan Lehnert1,4, Cristina Aguayo-Mazzucato5, Rui Cheng-Xue6, Patrick Gilon6, Iven Van Mechelen4, Susan Bonner-Weir5, Frédéric Lemaigre3, and Frans Schuit1,4,$ 1 Gene Expression Unit, Dept. Molecular Cell Biology, Katholieke Universiteit Leuven, 3000 Leuven, Belgium 2 ESAT-SCD, Department of Electrical Engineering, Katholieke Universiteit Leuven, 3000 Leuven, Belgium 3 Université Catholique de Louvain, de Duve Institute, 1200 Brussels, Belgium 4 Center for Computational Systems Biology, Katholieke Universiteit Leuven, 3000 Leuven, Belgium 5 Section of Islet Transplantation and Cell Biology, Joslin Diabetes Center, Harvard University, Boston, MA 02215, US 6 Unité d’Endocrinologie et Métabolisme, University of Louvain Faculty of Medicine, 1200 Brussels, Belgium $ To whom correspondence should be addressed: Frans Schuit O&N1 Herestraat 49 - bus 901 3000 Leuven, Belgium Email: [email protected] Phone: +32 16 347227 , Fax: +32 16 345995 Running title: Disallowed genes Keywords: disallowance, tissue-specific, tissue maturation, gene expression, intersection-union test Abbreviations: UTR UnTranslated Region H3K27me3 Histone H3 trimethylation at lysine 27 H3K4me3 Histone H3 trimethylation at lysine 4 H3K9ac Histone H3 acetylation at lysine 9 BMEL Bipotential Mouse Embryonic Liver Downloaded from genome.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract We report on a hitherto poorly characterized class of genes which are expressed in all tissues, except in one. -

An Integrated Approach to Comprehensively Map the Molecular Context of Proteins
bioRxiv preprint doi: https://doi.org/10.1101/264788; this version posted February 13, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Liu et al. An integrated approach to comprehensively map the molecular context of proteins Xiaonan Liu1,2, Kari Salokas1,2, Fitsum Tamene1,2,3 and Markku Varjosalo1,2,3* 1Institute of Biotechnology, University of Helsinki, Helsinki 00014, Finland 2Helsinki Institute of Life Science, University of Helsinki, Helsinki 00014, Finland 3Proteomics Unit, University of Helsinki, Helsinki 00014, Finland *Corresponding author Abstract: Protein-protein interactions underlie almost all cellular functions. The comprehensive mapping of these complex cellular networks of stable and transient associations has been made available by affi nity purifi cation mass spectrometry (AP-MS) and more recently by proximity based labelling methods such as BioID. Due the advancements in both methods and MS instrumentation, an in-depth analysis of the whole human proteome is at grasps. In order to facilitate this, we designed and optimized an integrated approach utilizing MAC-tag combining both AP-MS and BioID in a single construct. We systematically applied this approach to 18 subcellular localization markers and generated a molecular context database, which can be used to defi ne molecular locations for any protein of interest. In addition, we show that by combining the AP-MS and BioID results we can also obtain interaction distances within a complex. Taken together, our combined strategy off ers comprehensive approach for mapping physical and functional protein interactions. Introduction: Majority of proteins do not function in isolation geting the endogenous bait protein, allowing and their interactions with other proteins defi ne purifi cation of the bait protein together with the their cellular functions. -

Inflammatory and Adipogenic Stromal Subpopulations in Visceral Adipose Tissue Of
RESEARCH COMMUNICATION Identification of functionally distinct fibro- inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice Chelsea Hepler1†, Bo Shan1, Qianbin Zhang1, Gervaise H Henry2, Mengle Shao1, Lavanya Vishvanath1, Alexandra L Ghaben1, Angela B Mobley3, Douglas Strand2, Gary C Hon4, Rana K Gupta1* 1Touchstone Diabetes Center, Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, United States; 2Department of Urology, University of Texas Southwestern Medical Center, Dallas, United States; 3Department of Immunology, University of Texas Southwestern Medical Center, Dallas, United States; 4Cecil H. and Ida Green Center for Reproductive Biology Sciences, Division of Basic Reproductive Biology Research, Department of Obstetrics and Gynecology, University of Texas Southwestern Medical Center, Dallas, United States Abstract White adipose tissue (WAT) remodeling is dictated by coordinated interactions between adipocytes and resident stromal-vascular cells; however, the functional heterogeneity of adipose stromal cells has remained unresolved. We combined single-cell RNA-sequencing and FACS to identify and isolate functionally distinct subpopulations of PDGFRb+ stromal cells within visceral WAT of adult mice. LY6C- CD9- PDGFRb+ cells represent highly adipogenic visceral adipocyte precursor cells (‘APCs’), whereas LY6C+ PDGFRb+ cells represent fibro-inflammatory progenitors (‘FIPs’). FIPs lack adipogenic capacity, display pro-fibrogenic/pro-inflammatory *For correspondence: -

Supplemental Figures 04 12 2017
Jung et al. 1 SUPPLEMENTAL FIGURES 2 3 Supplemental Figure 1. Clinical relevance of natural product methyltransferases (NPMTs) in brain disorders. (A) 4 Table summarizing characteristics of 11 NPMTs using data derived from the TCGA GBM and Rembrandt datasets for 5 relative expression levels and survival. In addition, published studies of the 11 NPMTs are summarized. (B) The 1 Jung et al. 6 expression levels of 10 NPMTs in glioblastoma versus non‐tumor brain are displayed in a heatmap, ranked by 7 significance and expression levels. *, p<0.05; **, p<0.01; ***, p<0.001. 8 2 Jung et al. 9 10 Supplemental Figure 2. Anatomical distribution of methyltransferase and metabolic signatures within 11 glioblastomas. The Ivy GAP dataset was downloaded and interrogated by histological structure for NNMT, NAMPT, 12 DNMT mRNA expression and selected gene expression signatures. The results are displayed on a heatmap. The 13 sample size of each histological region as indicated on the figure. 14 3 Jung et al. 15 16 Supplemental Figure 3. Altered expression of nicotinamide and nicotinate metabolism‐related enzymes in 17 glioblastoma. (A) Heatmap (fold change of expression) of whole 25 enzymes in the KEGG nicotinate and 18 nicotinamide metabolism gene set were analyzed in indicated glioblastoma expression datasets with Oncomine. 4 Jung et al. 19 Color bar intensity indicates percentile of fold change in glioblastoma relative to normal brain. (B) Nicotinamide and 20 nicotinate and methionine salvage pathways are displayed with the relative expression levels in glioblastoma 21 specimens in the TCGA GBM dataset indicated. 22 5 Jung et al. 23 24 Supplementary Figure 4.