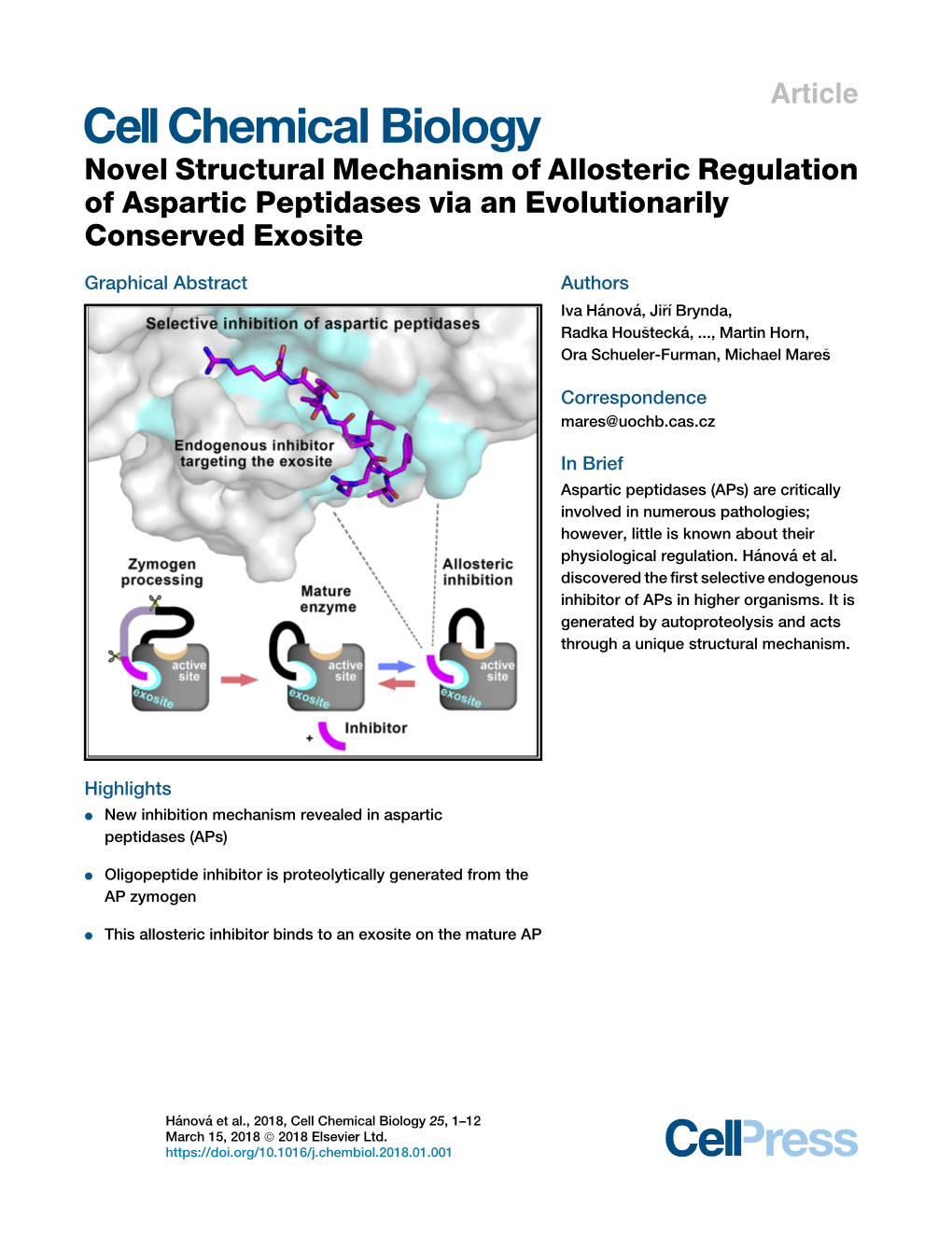
Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases Via an Evolutionarily Conserved Exosite
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chem331 Tansey Chpt4
3/10/20 Adhesion Covalent Cats - Proteases Roles Secretion P. gingivalis protease signal peptidases Immune Development Response T-cell protease matriptase 4 classes of proteases: Serine, Thiol (Cys), Acid Blood pressure Digestion regulation (Aspartyl), & Metal (Zn) trypsin renin Function Protease ex. Serine Cysteine trypsin, subtilisin, a-lytic Complement protease protease Nutrition Cell fusion protease Fixation hemaglutinase Invasion matrixmetalloproteases CI protease Evasion IgA protease Reproduction Tumor ADAM (a disintegrain and Invasion Adhesion collagenase and metalloproteinase) Fertilization signal peptidase, viral Processing acronase proteases, proteasome Signaling caspases, granzymes Fibrinolysis Pain Sensing tissue plasminogen kallikrein Acid Metallo- actvator protease protease Animal Virus Hormone Replication Processing HIV protease Kex 2 Substrate Specificity Serine Proteases Binding pocket is responsible for affinity Ser, His and Asp in active site Ser Proteases Multiple Mechanism Chymotrypsin Mechanism •Serine not generally an active amino acid for acid/base catalysis Nucleophilic attack •Catalytic triad of serine, histadine and aspartate responsible for the reactivity of serine in this active site •Covalent catalysis, Acid/base Catalysis, transition state bindingand Proximity mechanisms are used •Two phase reaction when an ester is used – burst phase - E +S initial reactions – steady state phase - EP -> E + P (deacylation) – The first step is the covalent catalysis - where the substrate actually is bound to the enzyme itself -

Elevated Calcium and Activation of Trypsinogen in Rat Pancreatic Acini Gut: First Published As 10.1136/Gut.41.3.339 on 1 September 1997
Gut 1997; 41: 339–343 339 Elevated calcium and activation of trypsinogen in rat pancreatic acini Gut: first published as 10.1136/gut.41.3.339 on 1 September 1997. Downloaded from T W Frick, C Fernández-del-Castillo, D Bimmler, A L Warshaw Abstract injection of calcium.5 Other experimental pro- Background—Acute pancreatitis associ- tocols used low dose continuous infusions of ated with hypercalcaemia has been de- calcium leading to a twofold increase in serum scribed in humans and experimental ionised calcium. Morphological changes of animals. It has been demonstrated that early acute pancreatitis were seen in several calcium dose dependently accelerates animal species.67 It was shown that hypercal- trypsinogen activation, and it is generally caemia induced a secretory block and accumu- believed that ectopic activation of diges- lation of digestive zymogens within the pancre- tive enzymes is an early event in the atic acinar cell.8 Zymogen activation, in pathophysiology of acute pancreatitis. particular trypsinogen, in homogenates of pan- Aims and methods—Trypsinogen activa- creatic tissue after calcium injection, suggested tion peptide (TAP) was measured in that the combination of zymogen accumulation isolated rat pancreatic acini exposed to and increased calcium leads to increased intra- elevated extracellular calcium in order to pancreatic trypsinogen activation as a very investigate the association between cal- early step in the pathogenesis of acute hyper- cium and trypsinogen activation in living calcaemia induced pancreatitis.58 cells. TAP was determined in the culture Despite this evidence it remained unclear medium either before (extracellular com- whether the ectopic zymogen activation oc- partment) or after (intracellular com- curred as an initial step in the pathogenesis of partment) cell homogenisation. -

DIGESTIVE SYSTEM -3 Emma Jakoi
Introductory Human Physiology ©copyright Emma Jakoi DIGESTIVE SYSTEM -3 Emma Jakoi. Ph.D. LEARNING OBJECTIVES 1. Explain the mechanisms of digestion and absorption of nutrients and identify where these occur within the gastrointestinal tube. 2. Explain the mechanisms of absorption of water and identify where this occurs within the gastrointestinal tube. 3. Explain the underlying mechanism for diarrhea and its causes. SMALL INTESTINE & NUTRIENT ABSORPTION Muscle contractions cause a ripple like movement that carries the food down the small intestine –like a conveyor belt. This transit is normally slow occurring over several hours. As complex food moves within the lumen of the small intestine, it is digested into small molecules. Subsequently these small molecules such as amino acids and sugars are absorbed into the body. These functions are coordinated by hormones. The small intestine is divided into three regions: duodenum, jejunum and ileum. The first, duodenum, is 10 inches long; the other two total 10 feet. The initial segment, the duodenum, receives the acidic chyme. Here the epithelium contains mucous glands and goblet cells which secrete mucus to neutralize the pH of the chyme. The duodenal epithelium cells also secrete hormones (Fig 1), cholecystokinin (CCK) and secretin, which signal the arrival of food to the pancreas, gall bladder, and stomach, respectively (Fig 1). Secretions from the pancreas and gall bladder are delivered directly to the lumen of the duodenum. Chyme G cells of stomach Duodenum CHO fats & peptides acid GLP-1 CCK Secretin Pancreas Pancreas Gall bladder Pancreas Islet Insulin enzymes bile salts HCO3- (Blood, feedforward) Figure 1. Digestive products signal the release of 2 hormones CCK and secretin from the duodenum and glucagon like peptide 1 (GLP-1) from the ileum. -

The Initiating Mechanism of Premature Trypsin Activation in Pancreatitis Kai Yang
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2004 The Initiating Mechanism of Premature Trypsin Activation in Pancreatitis Kai Yang Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] THE FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES THE INITIATING MECHANISM OF PREMATURE TRYPSIN ACTIVATION IN PANCREATITIS By KAI YANG A Thesis submitted to the Department of Biological Science in partial fulfillment of the requirements for the degree of Master of Science Degree Awarded: Summer Semester, 2004 The members of the Committee approve the Thesis of Kai Yang defended on 21 April, 2004. Wei-Chun Chin Professor Co - Directing Thesis George Bates Professor Co - Directing Thesis Thomas Keller Committee Member Laura Keller Committee Member Approved: Timothy S.Moerland, Chair, Department of Biological Science The Office of Graduate Studies has verified and approved the above named committee members. ii ACKNOWLEDGEMENTS I would like to express my thanks and appreciation to my advisor, Dr. Wei- Chun Chin for his continuous encouragement and kind help in my research and study. When I was in trouble, he gave me a chance to continue my study. I am also very grateful to Dr. George Bates, my co-major professor for his help. Without their support, I never would have finished my master degree in the department of biological science. I also wish to thank Dr. Thomas Keller and Dr. Laura Keller for their contribution as my committee members. I also would like to express my love and gratitude to my family for their support, patient, kindness, inspiration and love. -

Determination of Three Amino Acids Causing Alteration of Proteolytic Title Activities of Staphylococcal Glutamyl Endopeptidases
NAOSITE: Nagasaki University's Academic Output SITE Determination of three amino acids causing alteration of proteolytic Title activities of staphylococcal glutamyl endopeptidases. Nemoto, Takayuki K; Ono, Toshio; Shimoyama, Yu; Kimura, Shigenobu; Author(s) Ohara-Nemoto, Yuko Citation Biological chemistry, 390(3), pp.277-285; 2009 Issue Date 2009-03 URL http://hdl.handle.net/10069/23192 Right Walter de Gruyter. This document is downloaded at: 2020-09-17T22:23:39Z http://naosite.lb.nagasaki-u.ac.jp Determination of three amino acids that caused the alteration of proteolytic activities of staphylococcal glutamyl endopeptidases Takayuki K. Nemoto1, *, Toshio Ono1, Yu Shimoyama2, Shigenobu Kimura2 and Yuko Ohara-Nemoto1 1 Department of Oral Molecular Biology, Course of Medical and Dental Sciences, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki 852-8588 Japan 2 Department of Oral Microbiology, Iwate Medical University School of Dentistry, Morioka, 020-8505 Japan *Corresponding author T. K. Nemoto, Department of Oral Molecular Biology, Course of Medical and Dental Sciences, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki 852-8588, Japan Fax: +81 95 819 7640, Tel: +81 95 819 7642, E-mail: [email protected] Running title: Amino acid substitutions of V8 protease 1 Abstract Staphylococcus aureus, Staphylococcus epidermidis and Staphylococcus warneri secrete glutamyl endopeptidases, designated GluV8, GluSE and GluSW, respectively. The order of their protease activities was GluSE<GluSW<<GluV8. The present study investigated the mechanism that causes these differences. Expression of chimeric proteins between GluV8 and GluSE revealed that the difference was primarily attributed to amino acids at residues 170-195, which defined the intrinsic protease activity, and additionally to residues 119-169, which affected the proteolysis sensitivity. -

Apical Secretion of Lysosomal Enzymes in Rabbit Pancreas Occurs Via a Secretagogue Regulated Pathway and Is Increased After Pancreatic Duct Obstruction
Apical secretion of lysosomal enzymes in rabbit pancreas occurs via a secretagogue regulated pathway and is increased after pancreatic duct obstruction. T Hirano, … , M Saluja, M L Steer J Clin Invest. 1991;87(3):865-869. https://doi.org/10.1172/JCI115091. Research Article Lysosomal hydrolases such as cathepsin B are apically secreted from rabbit pancreatic acinar cells via a regulated as opposed to a constitutive pathway. Intravenous infusion of the cholecystokinin analogue caerulein results in highly correlated apical secretion of digestive and lysosomal enzymes, suggesting that they are discharged from the same presecretory compartment (zymogen granules). Lysosomal enzymes appear to enter that compartment as a result of missorting. After 7 h of duct obstruction is relieved, caerulein-stimulated apical secretion of cathepsin B and amylase is increased, but the ratio of cathepsin B to amylase secretion is not different than that following caerulein stimulation of animals never obstructed. These findings indicate that duct obstruction causes an increased amount of both lysosomal and digestive enzymes to accumulate within the secretagogue releasable compartment but that duct obstruction does not increase the degree of lysosomal enzyme missorting into that compartment. Pancreatic duct obstruction causes lysosomal hydrolases to become colocalized with digestive enzymes in organelles that, in size and distribution, resemble zymogen granules but that are not subject to secretion in response to secretagogue stimulation. These organelles may be of importance in the development of pancreatitis. Find the latest version: https://jci.me/115091/pdf Apical Secretion of Lysosomal Enzymes in Rabbit Pancreas Occurs Via a Secretagogue Regulated Pathway and Is Increased after Pancreatic Duct Obstruction T. -

Crystal Structure of the Zymogen Form of the Group a Streptococcus Virulence Factor Speb: an Integrin-Binding Cysteine Protease
Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: An integrin-binding cysteine protease Todd F. Kagawa*, Jakki C. Cooney†, Heather M. Baker*, Sean McSweeney‡, Mengyao Liu§¶, Siddeswar Gubba§, James M. Musser§¶, and Edward N. Baker*ʈ** *School of Biological Sciences and ʈDepartment of Chemistry, University of Auckland, Private Bag 92–019, Auckland, New Zealand; †Institute of Molecular Biosciences, Massey University, Palmerston North, New Zealand; ‡European Molecular Biology Laboratory Grenoble Outstation, 6 Av. J. Horowitz, Grenoble 38042, France; §Institute of Human Bacterial Pathogenesis, Department of Pathology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030; and ¶Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 903 South 4th Street, Hamilton, MT 59840 Communicated by Richard M. Krause, National Institutes of Health, Bethesda, MD, December 16, 1999 (received for review October 25, 1999) Pathogenic bacteria secrete protein toxins that weaken or disable The enzyme has fibrinolytic activity and causes myocardial their host, and thereby act as virulence factors. We have deter- necrosis when injected into rabbits (11). It also has been shown mined the crystal structure of streptococcal pyrogenic exotoxin B to trigger apoptosis (12), although the exact role of this process (SpeB), a cysteine protease that is a major virulence factor of the in pathogenesis is not known. Importantly, most strains of S. human pathogen Streptococcus pyogenes and participates in in- pyogenes that are associated with severe invasive disease express vasive disease episodes, including necrotizing fasciitis. The struc- a variant of SpeB that binds to integrins, which may be linked to ture, determined for the 40-kDa precursor form of SpeB at 1.6-Å their pathogenesis (13). -

Molecular Mechanism of Pancreatitis Dipendra Parajuli 01/05/02006
Molecular Mechanism of Pancreatitis Dipendra Parajuli 01/05/2006 University of Louisville Pancreatic Function in Health I. Inorganic Constituents - Aqueous, isotonic to plasma - Formed in duct cells - Consists of electrolytes and water Cations= Na+ , K+ Anions = Cl-, HCO3- - Electrolytes are secreted; H20 enters passively along the osmotic gradient - Proportion of Cl and HCO3 is dependent on the flow rate High Secretory rate HCO3 > Cl UniversityLow Secretory of rate Louisville HCO3 < Cl - Fluid is initially hypertonic but water movement into lumen dilutes it - Purposes of the water and ion secretions are to deliver digestive enzymes to the intestinal lumen and to help neutralize gastric acid emptied into duodenum - Impaired secretion of ions and water causes impaired flow of proteins causing thick secretions to plug the ducts University of Louisville Fluid secretion: Mechanism and Regulation - Export of H+ by Na+/H+ antiporter or H+-K+- ATPase -CO2 diffuses into cell - Carbonic anhydrase catalyzes HCO3 formation - - HCO3 transported out in exchange for Cl Cl- builds up in cell, exits by electrogenic channel =CFTR University of Louisville Fluid secretion: Mechanism and Regulation University of Louisville In Cystic Fibrosis - Defective CFTR (Cl- channel) - HCO3, Na+, water secretion impaired - Mucus buildup - maldigestion due to duct obstruction, loss of enzyme delivery to duodenum University of Louisville II. Organic Constituents Digestive Enzymes Proteolytic Enzymes Amyolytic Enzyme Lipolytic Enzymes Nucleases Trypsin Inhibitor Some of the enzymes are present in more than one form (e.g. trypsinogen =cationic, anionic and mesotrypsinogen ) University of Louisville Enzyme Secretion: Mechanism and Regulation Basic Secretory Unit = acinar cells. University of Louisville University of Louisville Acinar cells - Polarized Panuclear region - Rich in RER Apical region - Rich in Golgi apparatus and Zymogen granules University of Louisville Rough endoplasmic reticulum (RER). -

Zymogen Activation
Dorka Beri, Asst. Prof. In Biochemistry, GCW (A), Guntur Zymogen Activation A zymogen is like a wrapped candy bar. In order to get to the good stuff, you need to tear away what's keeping you from it. Zymogens, or proenzymes, are enzymes that aren't functioning yet because their action is blocked by a 'wrapper'. The 'wrapper' can be a link between two amino acids other enzymes are synthesized as inactive precursors that are subsequently activated by cleavage of one or a few specific peptide bonds. The inactive precursor is called a zymogen (or a proenzyme). A energy source (ATP) is not needed for cleavage. Therefore, in contrast with reversible regulation by phosphorylation, even proteins located outside cells can be activated by this means. You can recognize most zymogens by their name. Enzymes that begin with 'pro-' or end with '-gen' are often the zymogen form. For example: PROthrombin is the zymogen form of thrombin, an enzyme involved in blood clotting. PepsinoGEN is the zymogen form of pepsin, the enzyme found in your stomach that helps digest food. Why synthesized in inactive form : Zymogens also ensure the enzyme folds properly To prevent unwanted protein digestion make the enzyme stable in unfavorable environments, and allow the enzyme to go to the proper place so that it doesn't function where it's not supposed to. How they are activated : Dorka Beri, Asst. Prof. In Biochemistry, GCW (A), Guntur When the bonds are cut, the enzyme changes its conformation, so that the active site is free or able to become active. -

Activation Mechanisms for Zymogens Belonging to the Papain Family of Cysteine Proteases Were Investigated
ACTIVATION MECHANISMS FOR ZYMOGENS BELONGING TO THE PAPAIN FAMILY OF CYSTEINE PROTEASES OMAR QURAISHI Biochemistry Department McGill University, Montreal September 1999 A thesis submitted to the Faculty of Graduate Studies and Research in partial fulfilment of the requirements of the degree of Doctor of Philosophy O Omar Quraishi, 1999 National Library Bibliothèque nationale I*I of Canada du Canada Acquisitions and Acquisitions et Bibliographie Services services bibliographiques 395 Wellington Street 395, rue Wellington Ottawa ON KIA ON4 Ottawa ON KIA ON4 Canada Canada Yow file Votre rélérence Our fi& Notre rdférence The author has granted a non- L'auteur a accordé une licence non exclusive licence allowing the exclusive permettant à la National Library of Canada to Bibliothèque nationale du Canada de reproduce, loan, distribute or sell reproduire, prêter, distibuer ou copies of this thesis in microfom, vendre des copies de cette thèse sous paper or electronic formats. la forme de microfiche/fXm, de reproduction sur papier ou sur format électronique. The author retains ownership of the L'auteur conserve la propriété du copyright in this thesis. Neither the droit d'auteur qui protège cette thèse. thesis nor substantial extracts fiom it Ni la thèse ni des extraits substantiels may be printed or otherwise de celle-ci ne doivent être imprimés reproduced without the author's ou autrement reproduits sans son permission. autorisation. PAGE Acknowiedgments Abstract Résumé Introduction and Literature Review - Serine Proteases - Activation of Trypsinogen -

The Submembranous Network of Zymogen Granules 2235
Journal of Cell Science 113, 2233-2242 (2000) 2233 Printed in Great Britain © The Company of Biologists Limited 2000 JCS1273 A submembranous matrix of proteoglycans on zymogen granule membranes is involved in granule formation in rat pancreatic acinar cells Katja Schmidt1, Heidrun Dartsch1, Dietmar Linder2, Horst-Franz Kern1,* and Ralf Kleene1 1Institut für Zytobiologie und Zytopathologie, Philipps Universität, Robert-Koch-Str. 5, D-35033 Marburg, Germany 2Institut für Biochemie, Justus Liebig Universität, Gießen, Germany *Author for correspondence (e-mail: [email protected]) Accepted 27 March; published on WWW 25 May 2000 SUMMARY The secretory lectin ZG16p mediated the binding of antibody directed against chondroitin sulfate, indicating that aggregated zymogens to the granule membrane in the proteins represented proteoglycans. A staining pattern pancreatic acinar cells. Using a recently established in similar to the glycan reaction was observed in immunoblots vitro condensation-sorting assay, we now show that using a polyclonal antibody directed against the whole pretreatment of zymogen granule membranes (ZGM) with bicarbonate extract. Immunogold electron microscopy either sodium bicarbonate at pH 10 or with phosphatidyl revealed that this antibody reacted with components in the inositol-specific phospholipase C (PI-PLC) reduced the periphery of zymogen granules and strongly stained ZGM binding efficiency of zymogens to the same extent, as distinct in the pellet fraction of a standard in vitro condensation- components were liberated from ZGM. Analysis of the sorting assay. The amino acid composition of isolated composition of the bicarbonate extract revealed the presence components of both the acidic and basic group showed of the secretory lectin ZG16p, the serpin ZG46p and the similarities to aggrecan, a cartilage-specific proteoglycan, GPI-linked glycoprotein GP-2, together with several and to glycine-rich glycoproteins, respectively. -

Propeptide Processing and Proteolytic Activity of Proenzymes of the Staphylococcal and Enterococcal Gluv8-Family Protease
Indian Journal of Biochemistry & Biophysics Vol. 48, December 2012, pp. 421-427 Propeptide processing and proteolytic activity of proenzymes of the Staphylococcal and Enterococcal GluV8-family protease S M A Roufa, Y Ohara-Nemotoa, Y Shimoyamab, S Kimurab, T Onoa and T K Nemotoa* aDepartment of Oral Molecular Biology, Course of Medical and Dental Sciences, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki 852-8588, Japan bDivision of Molecular Microbiology, Iwate Medical University, Yahaba-cho 028-3694, Japan Received 21 January 2012; revised 30 August 2012 Proenzymes with various lengths of propeptides have been observed in GluV8 from Staphylococcus aureus and GluSE from S. epidermidis. However, the production mechanism of these proenzymes and roles of truncated propeptides have yet to be elucidated. Here we demonstrate that shortening of propeptide commonly occurs in an auto-catalytic manner in GluV8-family members, including those from coagulase negative Staphylococci and Enterococcus faecalis. Accompanied with propeptide shortening, the pro-mature junction (Asn/Ser-1-Val1) becomes more susceptible towards the hetero-catalytic maturation enzymes. The auto-catalytic propeptide truncation is not observed in Ser169Ala inert molecules of GluV8-family members. A faint proteolytic activity of proenzymes from Staphylococcus caprae and E. faecalis is detected. In addition, proteolytic activity of proenzyme of GluV8 carrying Arg-3AlaAsn-1 is demonstrated with synthetic peptide substrates LLE/Q-MCA. These results suggest that GluV8-family proenzymes with shortened propeptides intrinsically possess proteolytic activity and are involved in the propeptide shortening that facilitates the final hetero-catalytic maturation. Keywords: Enterococcus faecalis, GluV8, Propeptide, Staphylococcus aureus GluV8 from Staphylococcus aureus is produced as a observations suggest that mature region in pGluV8 zymogen (pGluV8) composed of an N-terminal is properly folded, independent of the structure of propeptide (Leu-39-Asn-1) and a mature portion propeptide.