Foamy-Like Endogenous Retroviruses Are Abundant and Extensive in Teleosts
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Non-Primate Lentiviral Vectors and Their Applications in Gene Therapy for Ocular Disorders
viruses Review Non-Primate Lentiviral Vectors and Their Applications in Gene Therapy for Ocular Disorders Vincenzo Cavalieri 1,2,* ID , Elena Baiamonte 3 and Melania Lo Iacono 3 1 Department of Biological, Chemical and Pharmaceutical Sciences and Technologies (STEBICEF), University of Palermo, Viale delle Scienze Edificio 16, 90128 Palermo, Italy 2 Advanced Technologies Network (ATeN) Center, University of Palermo, Viale delle Scienze Edificio 18, 90128 Palermo, Italy 3 Campus of Haematology Franco e Piera Cutino, Villa Sofia-Cervello Hospital, 90146 Palermo, Italy; [email protected] (E.B.); [email protected] (M.L.I.) * Correspondence: [email protected] Received: 30 April 2018; Accepted: 7 June 2018; Published: 9 June 2018 Abstract: Lentiviruses have a number of molecular features in common, starting with the ability to integrate their genetic material into the genome of non-dividing infected cells. A peculiar property of non-primate lentiviruses consists in their incapability to infect and induce diseases in humans, thus providing the main rationale for deriving biologically safe lentiviral vectors for gene therapy applications. In this review, we first give an overview of non-primate lentiviruses, highlighting their common and distinctive molecular characteristics together with key concepts in the molecular biology of lentiviruses. We next examine the bioengineering strategies leading to the conversion of lentiviruses into recombinant lentiviral vectors, discussing their potential clinical applications in ophthalmological research. Finally, we highlight the invaluable role of animal organisms, including the emerging zebrafish model, in ocular gene therapy based on non-primate lentiviral vectors and in ophthalmology research and vision science in general. Keywords: FIV; EIAV; BIV; JDV; VMV; CAEV; lentiviral vector; gene therapy; ophthalmology; zebrafish 1. -

Article Evolutionary Dynamics of the OR Gene Repertoire in Teleost Fishes
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.09.434524; this version posted March 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Article Evolutionary dynamics of the OR gene repertoire in teleost fishes: evidence of an association with changes in olfactory epithelium shape Maxime Policarpo1, Katherine E Bemis2, James C Tyler3, Cushla J Metcalfe4, Patrick Laurenti5, Jean-Christophe Sandoz1, Sylvie Rétaux6 and Didier Casane*,1,7 1 Université Paris-Saclay, CNRS, IRD, UMR Évolution, Génomes, Comportement et Écologie, 91198, Gif-sur-Yvette, France. 2 NOAA National Systematics Laboratory, National Museum of Natural History, Smithsonian Institution, Washington, D.C. 20560, U.S.A. 3Department of Paleobiology, National Museum of Natural History, Smithsonian Institution, Washington, D.C., 20560, U.S.A. 4 Independent Researcher, PO Box 21, Nambour QLD 4560, Australia. 5 Université de Paris, Laboratoire Interdisciplinaire des Energies de Demain, Paris, France 6 Université Paris-Saclay, CNRS, Institut des Neurosciences Paris-Saclay, 91190, Gif-sur- Yvette, France. 7 Université de Paris, UFR Sciences du Vivant, F-75013 Paris, France. * Corresponding author: e-mail: [email protected]. !1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.09.434524; this version posted March 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Teleost fishes perceive their environment through a range of sensory modalities, among which olfaction often plays an important role. -

Guide for Common Viral Diseases of Animals in Louisiana
Sampling and Testing Guide for Common Viral Diseases of Animals in Louisiana Please click on the species of interest: Cattle Deer and Small Ruminants The Louisiana Animal Swine Disease Diagnostic Horses Laboratory Dogs A service unit of the LSU School of Veterinary Medicine Adapted from Murphy, F.A., et al, Veterinary Virology, 3rd ed. Cats Academic Press, 1999. Compiled by Rob Poston Multi-species: Rabiesvirus DCN LADDL Guide for Common Viral Diseases v. B2 1 Cattle Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 2 Deer and Small Ruminants Please click on the principle system involvement Generalized viral disease Respiratory viral disease Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 3 Swine Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 4 Horses Please click on the principle system involvement Generalized viral diseases Neurological viral diseases Respiratory viral diseases Enteric viral diseases Abortifacient/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 5 Dogs Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. -

Fao/Government Cooperative Programme Scientific Basis
FI:GCP/RLA/140/JPN TECHNICAL DOCUMENT No. 4 FAO/GOVERNMENT COOPERATIVE PROGRAMME SCIENTIFIC BASIS FOR ECOSYSTEM-BASED MANAGEMENT IN THE LESSER ANTILLES INCLUDING INTERACTIONS WITH MARINE MAMMALS AND OTHER TOP PREDATORS CRUISE REPORT FOR THE LAPE ECOSYSTEM SURVEY ON RV CELTIC EXPLORER (CE0607) FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS Barbados, 2006 FI:GCP/RLA/140/JPN TECHNICAL DOCUMENT No. 4 FAO/GOVERNMENT COOPERATIVE PROGRAMME SCIENTIFIC BASIS FOR ECOSYSTEM-BASED MANAGEMENT IN THE LESSER ANTILLES INCLUDING INTERACTIONS WITH MARINE MAMMALS AND OTHER TOP PREDATORS CRUISE REPORT FOR THE LAPE ECOSYSTEM SURVEY ON RV CELTIC EXPLORER (CE0607) Lesser Antilles Pelagic Ecosystem Project (GCP/RLA/140/JPN) Bridgetown, Barbados FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS Barbados, 2006 This technical report is one of a series of reports prepared during the course of the project identified on the title page. The conclusions and recommendations given in the report are those considered appropriate at the time of its preparation. They may be modified in the light of further knowledge gained at subsequent stages of the project. The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations concerning the legal or development status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries All rights reserved. Reproduction and dissemination of material in this information product for educational or other non-commercial purposes are authorized without any prior written permission from the copyright holders provided the source is fully acknowledged. -

Early Stages of Fishes in the Western North Atlantic Ocean Volume
ISBN 0-9689167-4-x Early Stages of Fishes in the Western North Atlantic Ocean (Davis Strait, Southern Greenland and Flemish Cap to Cape Hatteras) Volume One Acipenseriformes through Syngnathiformes Michael P. Fahay ii Early Stages of Fishes in the Western North Atlantic Ocean iii Dedication This monograph is dedicated to those highly skilled larval fish illustrators whose talents and efforts have greatly facilitated the study of fish ontogeny. The works of many of those fine illustrators grace these pages. iv Early Stages of Fishes in the Western North Atlantic Ocean v Preface The contents of this monograph are a revision and update of an earlier atlas describing the eggs and larvae of western Atlantic marine fishes occurring between the Scotian Shelf and Cape Hatteras, North Carolina (Fahay, 1983). The three-fold increase in the total num- ber of species covered in the current compilation is the result of both a larger study area and a recent increase in published ontogenetic studies of fishes by many authors and students of the morphology of early stages of marine fishes. It is a tribute to the efforts of those authors that the ontogeny of greater than 70% of species known from the western North Atlantic Ocean is now well described. Michael Fahay 241 Sabino Road West Bath, Maine 04530 U.S.A. vi Acknowledgements I greatly appreciate the help provided by a number of very knowledgeable friends and colleagues dur- ing the preparation of this monograph. Jon Hare undertook a painstakingly critical review of the entire monograph, corrected omissions, inconsistencies, and errors of fact, and made suggestions which markedly improved its organization and presentation. -

Índice General
INSTITUTO POLITÉCNICO NACIONAL CENTRO INTERDISCIPLINARIO DE CIENCIAS MARINAS DEPARTAMENTO DE PESQUERÍAS Y BIOLOGÍA MARINA PATRONES LATITUDINALES DE COMPOSICIÓN Y DIVERSIDAD FUNCIONAL DE PECES ASOCIADOS A LA PESCA DE CAMARÓN DEL PACÍFICO MEXICANO TESIS QUE PARA OBTENER EL GRADO DE DOCTOR EN CIENCIAS MARINAS PRESENTA DEIVIS SAMUEL PALACIOS SALGADO LA PAZ, B.C.S., JUNIO DE 2011 SIP-14 BIS .INSTITUTO POLITÉCNICO NACIONAL SECRETARIA DE INVESTIGACIÓN Y POSGRADO ACTA DE REVISIÓN DE TESIS En la Ciudad de La Paz, B.C.S., siendo las 12:00 horas del día 23 del mes de Mayo del 2011 se reunieron los miembros de la Comisión Revisora de Tesis designada por el Colegio de Profesores de Estudios de Posgrado e Investigación de CICIMAR para examinar la tesis titulada: "PATRONES LATITUDINALES DE COMPOSICIÓN Y DIVERSIDAD FUNCIONAL DE PECES ASOCIADOS A LA PESCA DE CAMARÓN DEL PACÍFICO MEXICANO" Presentada por el alumno: PALACIOS SALDADO DEIVIS SAMUEL Apellido paterno materno nombre(s) Con registro: o 7 2 1 9 Aspirante de: DOCTORADO EN CIENCIAS MARINAS Después de intercambiar opiniones los miembros de la Comisión manifestaron APROBAR LA DEFENSA DE LA TESIS, en virtud de que satisface los requisitos señalados por las disposiciones reglamentarias vigentes. LA COMISIÓN REVISORA Directores de Tesis DR. MAN TINA REJÓN or de DR. FRANCISCO ARREGUIN SÁ DR. LEONARDO ANDR lA CÁRDENAS DR. HORACIO PÉREZ ESPAÑA PRESIDENTE DEL COLEGIO DE PROFESORES IPN CICIMAR DIRECCIÓN INSTITUTO POLITÉCNICO NACIONAL SECRETARÍA DE INVESTIGACIÓN Y POSGRADO CARTA CESIÓN DE DERECHOS En la Ciudad de La Paz, B.C.S., el día 01 del mes Junio del año 2011 el (la) que suscribe MC. -

The Expression of Human Endogenous Retroviruses Is Modulated by the Tat Protein of HIV‐1
The Expression of Human Endogenous Retroviruses is modulated by the Tat protein of HIV‐1 by Marta Jeannette Gonzalez‐Hernandez A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Immunology) in The University of Michigan 2012 Doctoral Committee Professor David M. Markovitz, Chair Professor Gary Huffnagle Professor Michael J. Imperiale Associate Professor David J. Miller Assistant Professor Akira Ono Assistant Professor Christiane E. Wobus © Marta Jeannette Gonzalez‐Hernandez 2012 For my family and friends, the most fantastic teachers I have ever had. ii Acknowledgements First, and foremost, I would like to thank David Markovitz for his patience and his scientific and mentoring endeavor. My time in the laboratory has been an honor and a pleasure. Special thanks are also due to all the members of the Markovitz laboratory, past and present. It has been a privilege, and a lot of fun, to work near such excellent scientists and friends. You all have a special place in my heart. I would like to thank all the members of my thesis committee for all the valuable advice, help and jokes whenever needed. Our collaborators from the Bioinformatics Core, particularly James Cavalcoli, Fan Meng, Manhong Dai, Maureen Sartor and Gil Omenn gave generous support, technical expertise and scientific insight to a very important part of this project. Thank you. Thanks also go to Mariana Kaplan’s and Akira Ono’s laboratory for help with experimental designs and for being especially generous with time and reagents. iii Table of Contents Dedication ............................................................................................................................ ii Acknowledgements ............................................................................................................. iii List of Figures ................................................................................................................... -

And Giant Guitarfish (Rhynchobatus Djiddensis)
VIRAL DISCOVERY IN BLUEGILL SUNFISH (LEPOMIS MACROCHIRUS) AND GIANT GUITARFISH (RHYNCHOBATUS DJIDDENSIS) BY HISTOPATHOLOGY EVALUATION, METAGENOMIC ANALYSIS AND NEXT GENERATION SEQUENCING by JENNIFER ANNE DILL (Under the Direction of Alvin Camus) ABSTRACT The rapid growth of aquaculture production and international trade in live fish has led to the emergence of many new diseases. The introduction of novel disease agents can result in significant economic losses, as well as threats to vulnerable wild fish populations. Losses are often exacerbated by a lack of agent identification, delay in the development of diagnostic tools and poor knowledge of host range and susceptibility. Examples in bluegill sunfish (Lepomis macrochirus) and the giant guitarfish (Rhynchobatus djiddensis) will be discussed here. Bluegill are popular freshwater game fish, native to eastern North America, living in shallow lakes, ponds, and slow moving waterways. Bluegill experiencing epizootics of proliferative lip and skin lesions, characterized by epidermal hyperplasia, papillomas, and rarely squamous cell carcinoma, were investigated in two isolated poopulations. Next generation genomic sequencing revealed partial DNA sequences of an endogenous retrovirus and the entire circular genome of a novel hepadnavirus. Giant Guitarfish, a rajiform elasmobranch listed as ‘vulnerable’ on the IUCN Red List, are found in the tropical Western Indian Ocean. Proliferative skin lesions were observed on the ventrum and caudal fin of a juvenile male quarantined at a public aquarium following international shipment. Histologically, lesions consisted of papillomatous epidermal hyperplasia with myriad large, amphophilic, intranuclear inclusions. Deep sequencing and metagenomic analysis produced the complete genomes of two novel DNA viruses, a typical polyomavirus and a second unclassified virus with a 20 kb genome tentatively named Colossomavirus. -

Feline Foamy Virus-Based Vectors: Advantages of an Authentic Animal Model
Viruses 2013, 5, 1702-1718; doi:10.3390/v5071702 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review Feline Foamy Virus-Based Vectors: Advantages of an Authentic Animal Model †,‡ † Weibin Liu , Janet Lei , Yang Liu, Dragana Slavkovic Lukic, Ann-Mareen Räthe, # Qiuying Bao, Timo Kehl, Anne Bleiholder, Torsten Hechler and Martin Löchelt * Department of Genome Modifications, Research Program Infection and Cancer, German Cancer Research Center, Im Neuenheimer Feld 242, 69120 Heidelberg, Germany; E-Mails: [email protected] (W.L.); [email protected] (J.L.); [email protected] (Y.L.); [email protected] (D.S.L.); [email protected] (A.-M.R.); [email protected] (Q.B.); [email protected] (T.K.); [email protected] (T.H.) † These authors contributed equally to this work. ‡ Current address: Department of Biochemistry and Molecular Biology, Mayo Clinic College of Medicine, 200 First Street SW, Rochester, MN 55905, USA. # Current address: Department of Biochemistry, Heidelberg Pharma GmbH, Schriesheimer Str. 101, 68526 Ladenburg, Germany. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +49-6221-424933; Fax: +49-6221-424932. Received: 1 March 2013; in revised form: 13 June 2013 / Accepted: 25 June 2013 / Published: 12 July 2013 Abstract: New-generation retroviral vectors have potential applications in vaccination and gene therapy. Foamy viruses are particularly interesting as vectors, because they are not associated to any disease. Vector research is mainly based on primate foamy viruses (PFV), but cats are an alternative animal model, due to their smaller size and the existence of a cognate feline foamy virus (FFV). -

Study of Chikungunya Virus Entry and Host Response to Infection Marie Cresson
Study of chikungunya virus entry and host response to infection Marie Cresson To cite this version: Marie Cresson. Study of chikungunya virus entry and host response to infection. Virology. Uni- versité de Lyon; Institut Pasteur of Shanghai. Chinese Academy of Sciences, 2019. English. NNT : 2019LYSE1050. tel-03270900 HAL Id: tel-03270900 https://tel.archives-ouvertes.fr/tel-03270900 Submitted on 25 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. N°d’ordre NNT : 2019LYSE1050 THESE de DOCTORAT DE L’UNIVERSITE DE LYON opérée au sein de l’Université Claude Bernard Lyon 1 Ecole Doctorale N° 341 – E2M2 Evolution, Ecosystèmes, Microbiologie, Modélisation Spécialité de doctorat : Biologie Discipline : Virologie Soutenue publiquement le 15/04/2019, par : Marie Cresson Study of chikungunya virus entry and host response to infection Devant le jury composé de : Choumet Valérie - Chargée de recherche - Institut Pasteur Paris Rapporteure Meng Guangxun - Professeur - Institut Pasteur Shanghai Rapporteur Lozach Pierre-Yves - Chargé de recherche - CHU d'Heidelberg Rapporteur Kretz Carole - Professeure - Université Claude Bernard Lyon 1 Examinatrice Roques Pierre - Directeur de recherche - CEA Fontenay-aux-Roses Examinateur Maisse-Paradisi Carine - Chargée de recherche - INRA Directrice de thèse Lavillette Dimitri - Professeur - Institut Pasteur Shanghai Co-directeur de thèse 2 UNIVERSITE CLAUDE BERNARD - LYON 1 Président de l’Université M. -

Molecular Analysis of the Complete Genome of a Simian Foamy Virus Infecting Hylobates Pileatus (Pileated Gibbon) Reveals Ancient Co-Evolution with Lesser Apes
viruses Article Molecular Analysis of the Complete Genome of a Simian Foamy Virus Infecting Hylobates pileatus (pileated gibbon) Reveals Ancient Co-Evolution with Lesser Apes Anupama Shankar 1, Samuel D. Sibley 2, Tony L. Goldberg 2 and William M. Switzer 1,* 1 Laboratory Branch, Division of HIV/AIDS Prevention, Center for Disease Control and Prevention, Atlanta, GA 30329, USA 2 Department of Pathobiological Sciences, School of Veterinary Medicine, University of Wisconsin-Madison, Madison, WI 53706, USA * Correspondence: [email protected]; Tel.: +1-404-639-2019 Received: 1 April 2019; Accepted: 30 June 2019; Published: 3 July 2019 Abstract: Foamy viruses (FVs) are complex retroviruses present in many mammals, including nonhuman primates, where they are called simian foamy viruses (SFVs). SFVs can zoonotically infect humans, but very few complete SFV genomes are available, hampering the design of diagnostic assays. Gibbons are lesser apes widespread across Southeast Asia that can be infected with SFV, but only two partial SFV sequences are currently available. We used a metagenomics approach with next-generation sequencing of nucleic acid extracted from the cell culture of a blood specimen from a lesser ape, the pileated gibbon (Hylobates pileatus), to obtain the complete SFVhpi_SAM106 genome. We used Bayesian analysis to co-infer phylogenetic relationships and divergence dates. SFVhpi_SAM106 is ancestral to other ape SFVs with a divergence date of ~20.6 million years ago, reflecting ancient co-evolution of the host and SFVhpi_SAM106. Analysis of the complete SFVhpi_SAM106 genome shows that it has the same genetic architecture as other SFVs but has the longest recorded genome (13,885-nt) due to a longer long terminal repeat region (2,071 bp). -
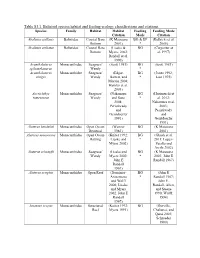
Table S3.1. Balistoid Species Habitat and Feeding Ecology Classifications and Citations
Table S3.1. Balistoid species habitat and feeding ecology classifications and citations. Species Family Habitat Habitat Feeding Feeding Mode Citation Mode Citation Abalistes stellaris Balistidae Coastal Bare (K Matsuura BG & EP (Kulbicki et al. Bottom 2001) * 2005) Abalistes stellatus Balistidae Coastal Bare (Lieske & BG (Carpenter et Bottom Myers, 2002; al. 1997) Randall et al. 1990) Acanthaluteres Monacanthidae Seagrass/ (Scott 1981) BG (Scott 1981) spilomelanurus Weedy * Acanthaluteres Monacanthidae Seagrass/ (Edgar, BG (Jones 1992; vittiger Weedy Barrett, and * Last 1975) Morton 2004; Hyndes et al. 2003) Acreichthys Monacanthidae Seagrass/ (Nakamura BG (Horinouchi et tomentosus Weedy and Sano * al. 2012; 2004; Nakamura et al. Peristiwady 2003; and Peristiwady Geistdoerfer and 1991) Geistdoerfer 1991) Aluterus heudeloti Monacanthidae Open Ocean (Wenner BG (K Matsuura Demersal 1983) 2001) Aluterus monoceros Monacanthidae Open Ocean (Kuiter 1992; BG (Ghosh et al. Rafting Lieske and * 2011; Lopez- Myers 2002) Peralta and Arcila 2002) Aluterus schoepfii Monacanthidae Seagrass/ (Lieske and BG (K Matsuura Weedy Myers 2002; * 2001; John E John E Randall 1967) Randall 1967) Aluterus scriptus Monacanthidae Open Reef (Dominici- BG (John E Arosemena * Randall 1967; and Wolff John E. 2006; Lieske Randall, Allen, and Myers and Steene 2002; John E 1990; Wulff, Randall 1994) 1967) Amanses scopas Monacanthidae Structured (Kuiter 1992; BG (Durville, Reef Myers 1991) Chabanet, and Quod 2003; Schroeder 1980) Balistapus Balistidae Structured (Hiatt and BG & EP (Hiatt and undulatus Reef Strasburg * Strasburg 1960; 1960; Lieske John E. Randall and Myers 1955; John E. 2002; Randall, Allen, McClanahan and Steene and Shafir 1990) 1990; John E. Randall, Allen, and Steene 1990) Balistes capriscus Balistidae Open Ocean (Longley BG & EP (Goldman, Pelagic 1941) * Glasgow, and Falk 2016; Lieske and Myers 2002; Vose and Nelson 1994) Balistes polylepis Balistidae Open Reef (Burgess et BG & EP (Abitia al.