Supplemental Material 1 (PDF)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterization of L-Serine Deaminases, Sdaa (PA2448) and Sdab 2 (PA5379), and Their Potential Role in Pseudomonas Aeruginosa 3 Pathogenesis 4 5 Sixto M
bioRxiv preprint doi: https://doi.org/10.1101/394957; this version posted August 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Characterization of L-serine deaminases, SdaA (PA2448) and SdaB 2 (PA5379), and their potential role in Pseudomonas aeruginosa 3 pathogenesis 4 5 Sixto M. Leal1,6, Elaine Newman2 and Kalai Mathee1,3,4,5 * 6 7 Author affiliations: 8 9 1Department of Biological Sciences, College of Arts Sciences and 10 Education, Florida International University, Miami, United States of 11 America 12 2Department of Biological Sciences, Concordia University, Montreal, 13 Canada 14 3Department of Molecular Microbiology and Infectious Diseases, Herbert 15 Wertheim College of Medicine, Florida International University, Miami, 16 United States of America 17 4Biomolecular Sciences Institute, Florida International University, Miami, 18 United States of America 19 20 Present address: 21 22 5Department of Human and Molecular Genetics, Herbert Wertheim 23 College of Medicine, Florida International University, Miami, United States 24 of America 25 6Case Western Reserve University, United States of America 26 27 28 *Correspondance: Kalai Mathee, MS, PhD, 29 [email protected] 30 31 Telephone : 1-305-348-0628 32 33 Keywords: Serine Catabolism, Central Metabolism, TCA Cycle, Pyruvate, 34 Leucine Responsive Regulatory Protein (LRP), One Carbon Metabolism 35 Running title: P. aeruginosa L-serine deaminases 36 Subject category: Pathogenicity and Virulence/Host Response 37 1 bioRxiv preprint doi: https://doi.org/10.1101/394957; this version posted August 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Bran Serine Hydrolases Achintya Kumar Dolui1,2, Arun Kumar Vijayakumar2,3, Ram Rajasekharan1,2,4 & Panneerselvam Vijayaraj1,2*
www.nature.com/scientificreports OPEN Activity‑based protein profling of rice (Oryza sativa L.) bran serine hydrolases Achintya Kumar Dolui1,2, Arun Kumar Vijayakumar2,3, Ram Rajasekharan1,2,4 & Panneerselvam Vijayaraj1,2* Rice bran is an underutilized agricultural by‑product with economic importance. The unique phytochemicals and fatty acid compositions of bran have been targeted for nutraceutical development. The endogenous lipases and hydrolases are responsible for the rapid deterioration of rice bran. Hence, we attempted to provide the frst comprehensive profling of active serine hydrolases (SHs) present in rice bran proteome by activity‑based protein profling (ABPP) strategy. The active site‑directed fuorophosphonate probe (rhodamine and biotin‑conjugated) was used for the detection and identifcation of active SHs. ABPP revealed 55 uncharacterized active‑SHs and are representing fve diferent known enzyme families. Based on motif and domain analyses, one of the uncharacterized and miss annotated SHs (Os12Ssp, storage protein) was selected for biochemical characterization by overexpressing in yeast. The purifed recombinant protein authenticated the serine protease activity in time and protein‑dependent studies. Os12Ssp exhibited the maximum activity at a pH between 7.0 and 8.0. The protease activity was inhibited by the covalent serine protease inhibitor, which suggests that the ABPP approach is indeed reliable than the sequence‑based annotations. Collectively, the comprehensive knowledge generated from this study would be useful in expanding the current understanding of rice bran SHs and paves the way for better utilization/ stabilization of rice bran. Rice (Oryza sativa) is one of the major staple foods for almost half the world’s population, especially in Asia 1. -

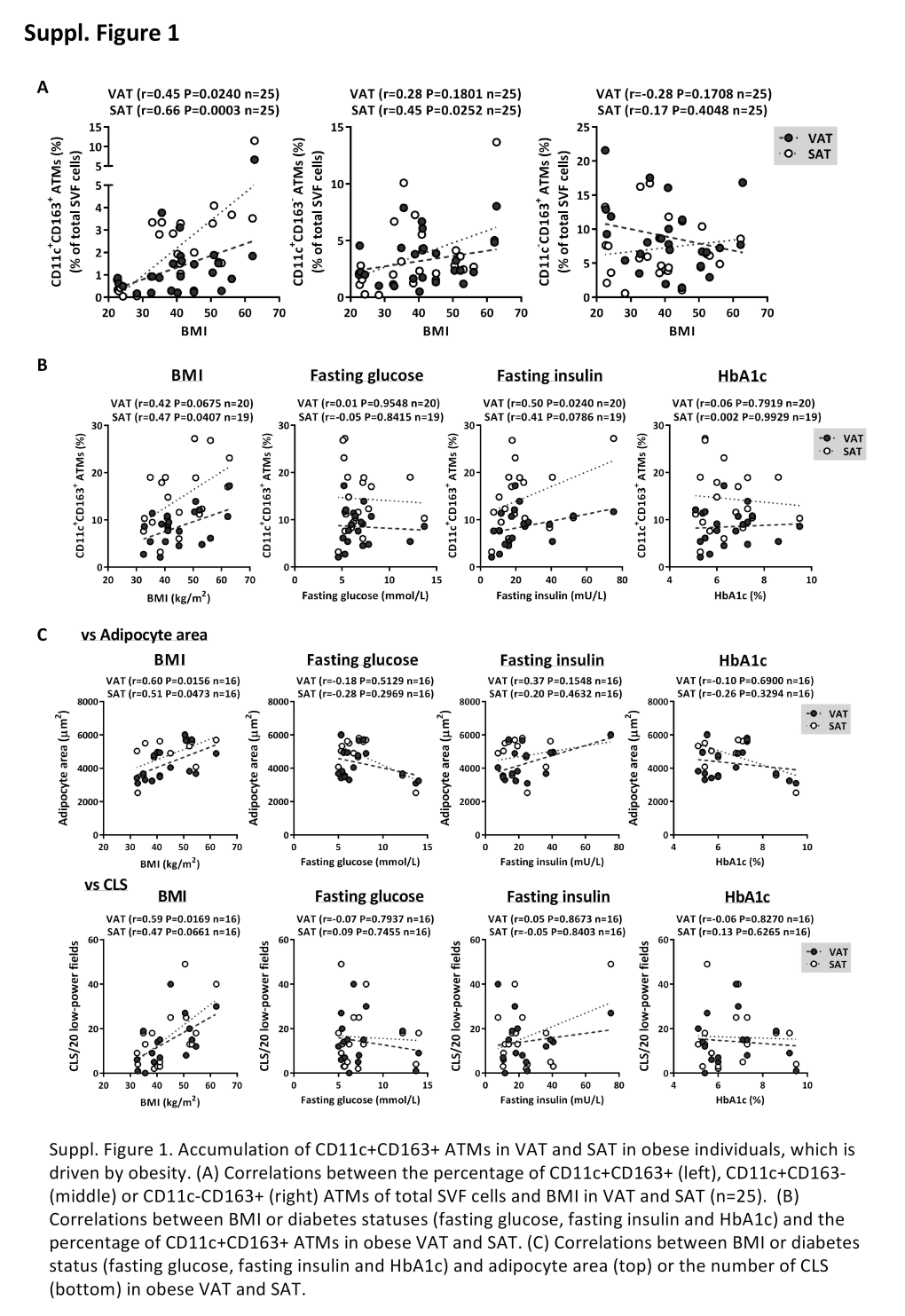
Supplemental Figure 1. Vimentin
Double mutant specific genes Transcript gene_assignment Gene Symbol RefSeq FDR Fold- FDR Fold- FDR Fold- ID (single vs. Change (double Change (double Change wt) (single vs. wt) (double vs. single) (double vs. wt) vs. wt) vs. single) 10485013 BC085239 // 1110051M20Rik // RIKEN cDNA 1110051M20 gene // 2 E1 // 228356 /// NM 1110051M20Ri BC085239 0.164013 -1.38517 0.0345128 -2.24228 0.154535 -1.61877 k 10358717 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 /// BC 1700025G04Rik NM_197990 0.142593 -1.37878 0.0212926 -3.13385 0.093068 -2.27291 10358713 NM_197990 // 1700025G04Rik // RIKEN cDNA 1700025G04 gene // 1 G2 // 69399 1700025G04Rik NM_197990 0.0655213 -1.71563 0.0222468 -2.32498 0.166843 -1.35517 10481312 NM_027283 // 1700026L06Rik // RIKEN cDNA 1700026L06 gene // 2 A3 // 69987 /// EN 1700026L06Rik NM_027283 0.0503754 -1.46385 0.0140999 -2.19537 0.0825609 -1.49972 10351465 BC150846 // 1700084C01Rik // RIKEN cDNA 1700084C01 gene // 1 H3 // 78465 /// NM_ 1700084C01Rik BC150846 0.107391 -1.5916 0.0385418 -2.05801 0.295457 -1.29305 10569654 AK007416 // 1810010D01Rik // RIKEN cDNA 1810010D01 gene // 7 F5 // 381935 /// XR 1810010D01Rik AK007416 0.145576 1.69432 0.0476957 2.51662 0.288571 1.48533 10508883 NM_001083916 // 1810019J16Rik // RIKEN cDNA 1810019J16 gene // 4 D2.3 // 69073 / 1810019J16Rik NM_001083916 0.0533206 1.57139 0.0145433 2.56417 0.0836674 1.63179 10585282 ENSMUST00000050829 // 2010007H06Rik // RIKEN cDNA 2010007H06 gene // --- // 6984 2010007H06Rik ENSMUST00000050829 0.129914 -1.71998 0.0434862 -2.51672 -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Role of Phospholipases in Adrenal Steroidogenesis
229 1 W B BOLLAG Phospholipases in adrenal 229:1 R29–R41 Review steroidogenesis Role of phospholipases in adrenal steroidogenesis Wendy B Bollag Correspondence should be addressed Charlie Norwood VA Medical Center, One Freedom Way, Augusta, GA, USA to W B Bollag Department of Physiology, Medical College of Georgia, Augusta University (formerly Georgia Regents Email University), Augusta, GA, USA [email protected] Abstract Phospholipases are lipid-metabolizing enzymes that hydrolyze phospholipids. In some Key Words cases, their activity results in remodeling of lipids and/or allows the synthesis of other f adrenal cortex lipids. In other cases, however, and of interest to the topic of adrenal steroidogenesis, f angiotensin phospholipases produce second messengers that modify the function of a cell. In this f intracellular signaling review, the enzymatic reactions, products, and effectors of three phospholipases, f phospholipids phospholipase C, phospholipase D, and phospholipase A2, are discussed. Although f signal transduction much data have been obtained concerning the role of phospholipases C and D in regulating adrenal steroid hormone production, there are still many gaps in our knowledge. Furthermore, little is known about the involvement of phospholipase A2, Endocrinology perhaps, in part, because this enzyme comprises a large family of related enzymes of that are differentially regulated and with different functions. This review presents the evidence supporting the role of each of these phospholipases in steroidogenesis in the Journal Journal of Endocrinology adrenal cortex. (2016) 229, R1–R13 Introduction associated GTP-binding protein exchanges a bound GDP for a GTP. The G protein with GTP bound can then Phospholipids serve a structural function in the cell in that activate the enzyme, phospholipase C (PLC), that cleaves they form the lipid bilayer that maintains cell integrity. -

Human Artificial Chromosome (Hac) Vector
Europäisches Patentamt *EP001559782A1* (19) European Patent Office Office européen des brevets (11) EP 1 559 782 A1 (12) EUROPEAN PATENT APPLICATION published in accordance with Art. 158(3) EPC (43) Date of publication: (51) Int Cl.7: C12N 15/09, C12N 1/15, 03.08.2005 Bulletin 2005/31 C12N 1/19, C12N 1/21, C12N 5/10, C12P 21/02 (21) Application number: 03751334.8 (86) International application number: (22) Date of filing: 03.10.2003 PCT/JP2003/012734 (87) International publication number: WO 2004/031385 (15.04.2004 Gazette 2004/16) (84) Designated Contracting States: • KATOH, Motonobu, Tottori University AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Yonago-shi, Tottori 683-8503 (JP) HU IE IT LI LU MC NL PT RO SE SI SK TR • TOMIZUKA, Kazuma, Designated Extension States: Kirin Beer Kabushiki Kaisha AL LT LV MK Takashi-shi, Gunma 370-1295 (JP) • KUROIWA, Yoshimi, (30) Priority: 04.10.2002 JP 2002292853 Kirin Beer Kabushiki Kaisha Takasaki-shi, Gunma 370-1295 (JP) (71) Applicant: KIRIN BEER KABUSHIKI KAISHA • KAKEDA, Minoru, Kirin Beer Kabushiki Kaisha Tokyo 104-8288 (JP) Takasaki-shi, Gunma 370-1295 (JP) (72) Inventors: (74) Representative: HOFFMANN - EITLE • OSHIMURA, Mitsuo, Tottori University Patent- und Rechtsanwälte Yonago-shi, Tottori 683-8503 (JP) Arabellastrasse 4 81925 München (DE) (54) HUMAN ARTIFICIAL CHROMOSOME (HAC) VECTOR (57) The present invention relates to a human arti- ing a cell which expresses foreign DNA. Furthermore, ficial chromosome (HAC) vector and a method for pro- the present invention relates to a method for producing ducing the same. -

An ER Stress/Defective Unfolded Protein Response Model Richard T
ORIGINAL RESEARCH Ethanol Induced Disordering of Pancreatic Acinar Cell Endoplasmic Reticulum: An ER Stress/Defective Unfolded Protein Response Model Richard T. Waldron,1,2 Hsin-Yuan Su,1 Honit Piplani,1 Joseph Capri,3 Whitaker Cohn,3 Julian P. Whitelegge,3 Kym F. Faull,3 Sugunadevi Sakkiah,1 Ravinder Abrol,1 Wei Yang,1 Bo Zhou,1 Michael R. Freeman,1,2 Stephen J. Pandol,1,2 and Aurelia Lugea1,2 1Department of Medicine, Cedars Sinai Medical Center, Los Angeles, California; 2Department of Medicine, or 3Psychiatry and Biobehavioral Sciences, University of California Los Angeles David Geffen School of Medicine, Los Angeles, California Pancreatic acinar cells Pancreatic acinar cells - no pathology - - Pathology - Ethanol feeding ER sXBP1 Pdi, Grp78… (adaptive UPR) aggregates Proper folding and secretion • disordered ER of proteins processed in the • impaired redox folding endoplasmic reticulum (ER) • ER protein aggregation • secretory defects SUMMARY METHODS: Wild-type and Xbp1þ/- mice were fed control and ethanol diets, then tissues were homogenized and fraction- Heavy alcohol consumption is associated with pancreas ated. ER proteins were labeled with a cysteine-reactive probe, damage, but light drinking shows the opposite effects, isotope-coded affinity tag to obtain a novel pancreatic redox ER reinforcing proteostasis through the unfolded protein proteome. Specific labeling of active serine hydrolases in ER with response orchestrated by X-box binding protein 1. Here, fluorophosphonate desthiobiotin also was characterized pro- ethanol-induced changes in endoplasmic reticulum protein teomically. Protein structural perturbation by redox changes redox and structure/function emerge from an unfolded was evaluated further in molecular dynamic simulations. protein response–deficient genetic model. -

HMGB1 in Health and Disease R
Donald and Barbara Zucker School of Medicine Journal Articles Academic Works 2014 HMGB1 in health and disease R. Kang R. C. Chen Q. H. Zhang W. Hou S. Wu See next page for additional authors Follow this and additional works at: https://academicworks.medicine.hofstra.edu/articles Part of the Emergency Medicine Commons Recommended Citation Kang R, Chen R, Zhang Q, Hou W, Wu S, Fan X, Yan Z, Sun X, Wang H, Tang D, . HMGB1 in health and disease. 2014 Jan 01; 40():Article 533 [ p.]. Available from: https://academicworks.medicine.hofstra.edu/articles/533. Free full text article. This Article is brought to you for free and open access by Donald and Barbara Zucker School of Medicine Academic Works. It has been accepted for inclusion in Journal Articles by an authorized administrator of Donald and Barbara Zucker School of Medicine Academic Works. Authors R. Kang, R. C. Chen, Q. H. Zhang, W. Hou, S. Wu, X. G. Fan, Z. W. Yan, X. F. Sun, H. C. Wang, D. L. Tang, and +8 additional authors This article is available at Donald and Barbara Zucker School of Medicine Academic Works: https://academicworks.medicine.hofstra.edu/articles/533 NIH Public Access Author Manuscript Mol Aspects Med. Author manuscript; available in PMC 2015 December 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Mol Aspects Med. 2014 December ; 0: 1–116. doi:10.1016/j.mam.2014.05.001. HMGB1 in Health and Disease Rui Kang1,*, Ruochan Chen1, Qiuhong Zhang1, Wen Hou1, Sha Wu1, Lizhi Cao2, Jin Huang3, Yan Yu2, Xue-gong Fan4, Zhengwen Yan1,5, Xiaofang Sun6, Haichao Wang7, Qingde Wang1, Allan Tsung1, Timothy R. -

Supplementary Figure S4
18DCIS 18IDC Supplementary FigureS4 22DCIS 22IDC C D B A E (0.77) (0.78) 16DCIS 14DCIS 28DCIS 16IDC 28IDC (0.43) (0.49) 0 ADAMTS12 (p.E1469K) 14IDC ERBB2, LASP1,CDK12( CCNE1 ( NUTM2B SDHC,FCGR2B,PBX1,TPR( CD1D, B4GALT3, BCL9, FLG,NUP21OL,TPM3,TDRD10,RIT1,LMNA,PRCC,NTRK1 0 ADAMTS16 (p.E67K) (0.67) (0.89) (0.54) 0 ARHGEF38 (p.P179Hfs*29) 0 ATG9B (p.P823S) (0.68) (1.0) ARID5B, CCDC6 CCNE1, TSHZ3,CEP89 CREB3L2,TRIM24 BRAF, EGFR (7p11); 0 ABRACL (p.R35H) 0 CATSPER1 (p.P152H) 0 ADAMTS18 (p.Y799C) 19q12 0 CCDC88C (p.X1371_splice) (0) 0 ADRA1A (p.P327L) (10q22.3) 0 CCNF (p.D637N) −4 −2 −4 −2 0 AKAP4 (p.G454A) 0 CDYL (p.Y353Lfs*5) −4 −2 Log2 Ratio Log2 Ratio −4 −2 Log2 Ratio Log2 Ratio 0 2 4 0 2 4 0 ARID2 (p.R1068H) 0 COL27A1 (p.G646E) 0 2 4 0 2 4 2 EDRF1 (p.E521K) 0 ARPP21 (p.P791L) ) 0 DDX11 (p.E78K) 2 GPR101, p.A174V 0 ARPP21 (p.P791T) 0 DMGDH (p.W606C) 5 ANP32B, p.G237S 16IDC (Ploidy:2.01) 16DCIS (Ploidy:2.02) 14IDC (Ploidy:2.01) 14DCIS (Ploidy:2.9) -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 Log Ratio Log Ratio Log Ratio Log Ratio 12DCIS 0 ASPM (p.S222T) Log Ratio Log Ratio 0 FMN2 (p.G941A) 20 1 2 3 2 0 1 2 3 2 ERBB3 (p.D297Y) 2 0 1 2 3 20 1 2 3 0 ATRX (p.L1276I) 20 1 2 3 2 0 1 2 3 0 GALNT18 (p.F92L) 2 MAPK4, p.H147Y 0 GALNTL6 (p.E236K) 5 C11orf1, p.Y53C (10q21.2); 0 ATRX (p.R1401W) PIK3CA, p.H1047R 28IDC (Ploidy:2.0) 28DCIS (Ploidy:2.0) 22IDC (Ploidy:3.7) 22DCIS (Ploidy:4.1) 18IDC (Ploidy:3.9) 18DCIS (Ploidy:2.3) 17q12 0 HCFC1 (p.S2025C) 2 LCMT1 (p.S34A) 0 ATXN7L2 (p.X453_splice) SPEN, p.P677Lfs*13 CBFB 1 2 3 4 5 6 7 8 9 10 11 -

DNA Supercoiling with a Twist Edwin Kamau Louisiana State University and Agricultural and Mechanical College, [email protected]
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 2005 DNA supercoiling with a twist Edwin Kamau Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Recommended Citation Kamau, Edwin, "DNA supercoiling with a twist" (2005). LSU Doctoral Dissertations. 999. https://digitalcommons.lsu.edu/gradschool_dissertations/999 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. DNA SUPERCOILING WITH A TWIST A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Department of Biological Sciences By Edwin Kamau B.S. Horticulture, Egerton University, Kenya, 1996, May, 2005 DEDICATIONS I would like to dedicate this dissertation to my son, Ashford Kimani Kamau. Your age marks the life of my graduate school career. I have been a weekend dad, broke and with enormous financial responsibilities; it kills me to see how sometimes you cry your heart out on Sunday evening when our weekend time together is over. I miss you too every moment, you have encouraged me to be a better person, a better dad so that I can make you proud. At your tender age, I have learned a lot from you. We will soon spend all the time in the world we need together; travel, sports, music, pre-historic discoveries and adventures; the road is wide open. -

Isolation and Nucleotide Sequence of the Cdna for Rat Liver Serine
Proc. Natl. Acad. Sci. USA Vol. 85, pp. 5809-5813, August 1988 Biochemistry Isolation and nucleotide sequence of the cDNA for rat liver serine dehydratase mRNA and structures of the 5' and 3' flanking regions of the serine dehydratase gene (threonine dehydratase/hormonal regulation/consensus sequences) HIROFUMI OGAWA*t, DUNCAN A. MILLER*, TRACY DUNN*, YEU SU*, JAMES M. BURCHAMt, CARL PERAINOt, MOTOJI FUJIOKAt, KAY BABCOCK*, AND HENRY C. PITOT*§ *McArdle Laboratory for Cancer Research, The Medical School, University of Wisconsin, Madison, WI 53706; tDepartment of Biochemistry, Toyama Medical and Pharmaceutical University, Faculty of Medicine, Sugitani, Toyama 930-01, Japan; and tDivision of Biological and Medical Research, Argonne National Laboratory, Argonne, IL 60439 Communicated by Van R. Potter, April 15, 1988 (received for review December 29, 1987) ABSTRACT Rat serine dehydratase cDNA clones were determination of the exact size of DNA complementary to isolated from a Agtll cDNA library on the basis of their serine dehydratase mRNA was made by S1 nuclease and reactivity with monospecific immunoglobulin to the purified sequencing of genomic clones of the regions flanking the enzyme. Using the cDNA insert from a clone that encoded the gene. serine dehydratase subunit as a probe, additional clones were isolated from the same library by plaque hybridization. Nucle- otide sequence analysis of the largest clone obtained showed MATERIALS AND METHODS that it has 1444 base pairs with an open reading frame consisting of 1089 base pairs. The deduced amino acid sequence cDNA Cloning. A rat liver cDNA library constructed in contained sequences of several portions of the serine dehydra- Agtll phage (13) was screened for antibody-reactive plaques tase protein, as determined by Edman degradation. -

Fucosyltransferase Genes on Porcine Chromosome 6Q11 Are Closely Linked to the Blood Group Inhibitor (S) and Escherichia Coli F18 Receptor (ECF18R) Loci
Mammalian Genome 8, 736–741 (1997). © Springer-Verlag New York Inc. 1997 Two a(1,2) fucosyltransferase genes on porcine Chromosome 6q11 are closely linked to the blood group inhibitor (S) and Escherichia coli F18 receptor (ECF18R) loci E. Meijerink,1 R. Fries,1,*P.Vo¨geli,1 J. Masabanda,1 G. Wigger,1 C. Stricker,1 S. Neuenschwander,1 H.U. Bertschinger,2 G. Stranzinger1 1Institute of Animal Science, Swiss Federal Institute of Technology, ETH-Zentrum, CH-8092 Zurich, Switzerland 2Institute of Veterinary Bacteriology, University of Zurich, CH 8057 Zurich, Switzerland Received: 17 February 1997 / Accepted: 30 May 1997 Abstract. The Escherichia coli F18 receptor locus (ECF18R) has fimbriae F107, has been shown to be genetically controlled by the been genetically mapped to the halothane linkage group on porcine host and is inherited as a dominant trait (Bertschinger et al. 1993) Chromosome (Chr) 6. In an attempt to obtain candidate genes for with B being the susceptibility allele and b the resistance allele. this locus, we isolated 5 cosmids containing the a(1,2)fucosyl- The genetic locus for this E. coli F18 receptor (ECF18R) has been transferase genes FUT1, FUT2, and the pseudogene FUT2P from mapped to porcine Chr 6 (SSC6), based on its close linkage to the a porcine genomic library. Mapping by fluorescence in situ hy- S locus and other loci of the halothane (HAL) linkage group (Vo¨- bridization placed all these clones in band q11 of porcine Chr 6 geli et al. 1996). The epistatic S locus suppresses the phenotypic (SSC6q11). Sequence analysis of the cosmids resulted in the char- expression of the A-0 blood group system when being SsSs (Vo¨geli acterization of an open reading frame (ORF), 1098 bp in length, et al.