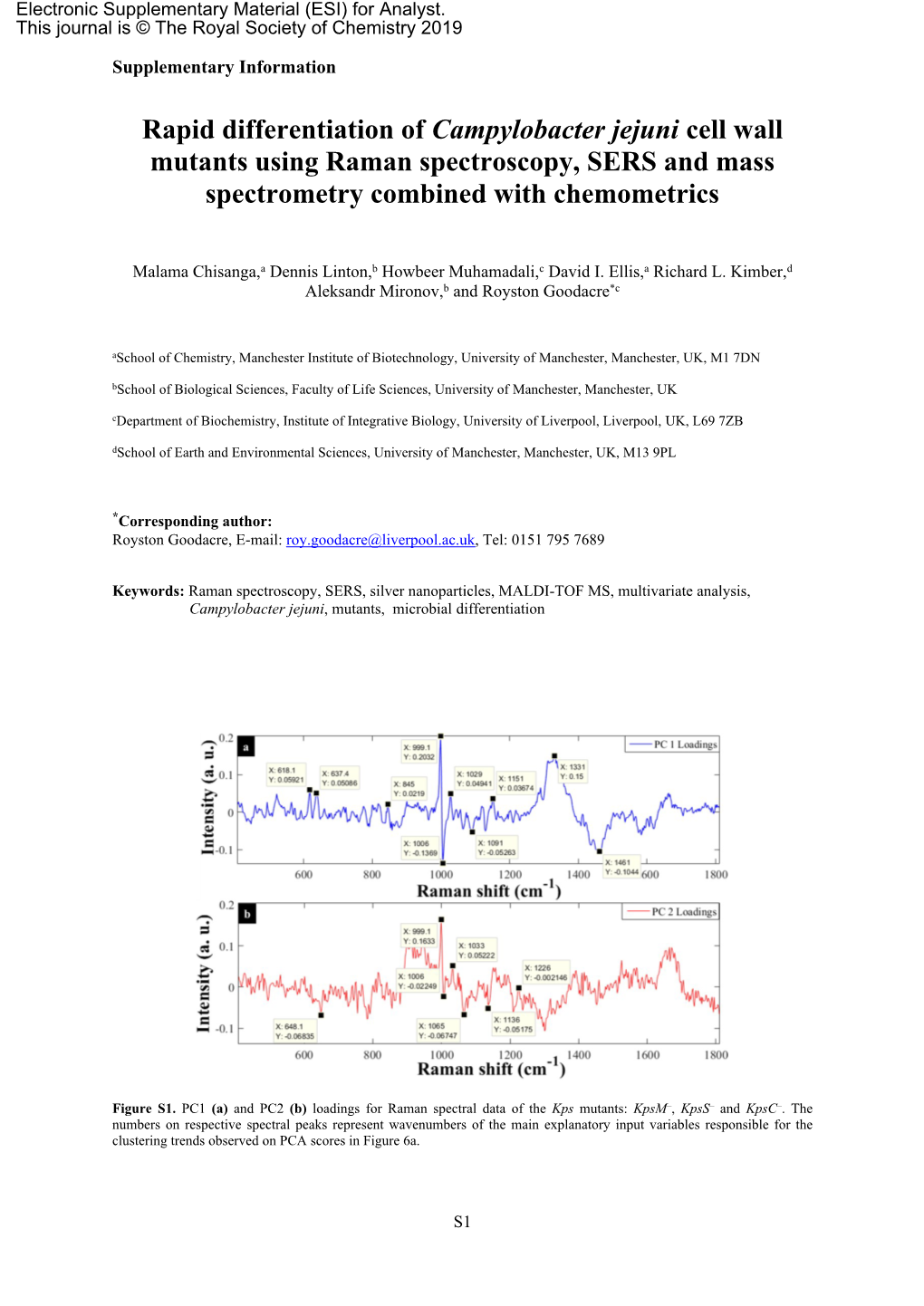
Supplementary Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Are Omics the Death of Good Sampling Practice?
VOL. 31 NO. 3 (2019) TONY DAVIES COLUMN Are omics the death of Good Sampling Practice? Antony N. Daviesa,b and Roy Goodacrec aExpert Capability Group – Measurement and Analytical Science, Nouryon, Deventer, the Netherlands bSERC, Sustainable Environment Research Centre, Faculty of Computing Engineering and Science, University of South Wales, UK. 0000-0002-3119-4202 cDepartment of Biochemistry, Institute of Integrative Biology, University of Liverpool, UK. E-mail: [email protected], 0000-0003-2230-645X During the recent Royal Society of apparently promising dead-ends. We are in most case-control studies the cases Chemistry, Faraday discussion meeting in reminded by George Poste in his edito- (those with some form of disease) are Edinburgh on Challenges in Analysis of rial “Bring on the Biomarkers” in 20112 usually already on medication (or self Complex Natural Mixtures I found myself that, at that time, of the 150,000 clini- medicating), so this strong confounding wondering if the power that our modern cal biomarkers described in the literature factor also needs to be considered. spectrometers bring to the study of highly a mere 100 were routinely used in the There is an enormous gap between complex systems can sometimes over- clinic. delivering theoretical correlations with whelm our natural scepticism around Omics experiments in themselves the hope of finding causation from stud- poor sampling practices.1 Some targeted present an enormous issue for classical ies of cell cultures in Petri dishes to questions put by Roy Goodacre in this statisticians just by their huge dimension- catching the developing lung cancer in a direction to several speakers seemed to ality. -

Rothamsted Repository Download
Patron: Her Majesty The Queen Rothamsted Research Harpenden, Herts, AL5 2JQ Telephone: +44 (0)1582 763133 WeB: http://www.rothamsted.ac.uk/ Rothamsted Repository Download A - Papers appearing in refereed journals Ward, J. L., Moing, A., Allwood, J. W., Aharoni, A., Baker, J., Beale, M. H., Ben-Dor, S., Biais, B., Brigante, F., Burger, Y., Deborde, C., Erban, A., Faigenboim, A., Gur, A., Goodacre, R., Hansen, T. H., Jacob, D., Katzir, N., Kopka, J., Lewinsohn, E., Maucourt, M., Meir, S., Miller, S., Mumm, R., Oren, E., Paris, H. S., Rogachev, I., Rolin, D., Saar, U., Schjoerring, J. K., Tadmor, Y., Tzuri, G., Vos, R. C. D., Ward, J. L., Yeselson, E., Hall, R. D. and Schaffer, A. A. 2020. Comparative Metabolomics and Molecular Phylogenetics of Melon (Cucumis melo, Cucurbitaceae) Biodiversity. Metabolites. 10 (3), pp. 121-147. The publisher's version can be accessed at: • https://dx.doi.org/10.3390/metabo10030121 • https://www.mdpi.com/2218-1989/10/3/121 The output can be accessed at: https://repository.rothamsted.ac.uk/item/9783y/comparative-metabolomics-and- molecular-phylogenetics-of-melon-cucumis-melo-cucurbitaceae-biodiversity. © 20 March 2020, Please contact [email protected] for copyright queries. 28/05/2020 09:19 repository.rothamsted.ac.uk [email protected] Rothamsted Research is a Company Limited by Guarantee Registered Office: as above. Registered in England No. 2393175. Registered Charity No. 802038. VAT No. 197 4201 51. Founded in 1843 by John Bennet Lawes. H OH metabolites OH Article Comparative Metabolomics and Molecular Phylogenetics of Melon (Cucumis melo, Cucurbitaceae) Biodiversity Annick Moing 1 , J. -

The Development of Enhanced Raman Scattering for the Trace Analysis of Biomolecules
The Development of Enhanced Raman Scattering for the Trace Analysis of Biomolecules A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy in the Faculty of Engineering and Physical Sciences 2013 David Paul Cowcher Supervised by Professor Royston Goodacre School of Chemistry Contents Contents CONTENTS ....................................................................................................................................... 2 LIST OF FIGURES .......................................................................................................................... 7 LIST OF TABLES .......................................................................................................................... 14 LIST OF EQUATIONS .................................................................................................................. 15 LIST OF SCHEMES ...................................................................................................................... 15 LIST OF ABBREVIATIONS ........................................................................................................ 16 ABSTRACT ..................................................................................................................................... 18 ACKNOWLEDGEMENTS ............................................................................................................ 19 DECLARATION ............................................................................................................................ -

Surface-Enhanced Raman Scattering (SERS)
Focal Point Review Applied Spectroscopy 2018, Vol. 72(7) 987–1000 ! The Author(s) 2018 Surface-Enhanced Raman Scattering Reprints and permissions: sagepub.co.uk/journalsPermissions.nav (SERS) in Microbiology: Illumination and DOI: 10.1177/0003702818764672 journals.sagepub.com/home/asp Enhancement of the Microbial World Malama Chisanga, Howbeer Muhamadali, David I. Ellis, and Royston Goodacre Abstract The microbial world forms a huge family of organisms that exhibit the greatest phylogenetic diversity on Earth and thus colonize virtually our entire planet. Due to this diversity and subsequent complex interactions, the vast majority of microorganisms are involved in innumerable natural bioprocesses and contribute an absolutely vital role toward the maintenance of life on Earth, whilst a small minority cause various infectious diseases. The ever-increasing demand for environmental monitoring, sustainable ecosystems, food security, and improved healthcare systems drives the continuous search for inexpensive but reproducible, automated and portable techniques for detection of microbial isolates and understanding their interactions for clinical, environmental, and industrial applications and benefits. Surface-enhanced Raman scattering (SERS) is attracting significant attention for the accurate identification, discrimination and characteriza- tion and functional assessment of microbial cells at the single cell level. In this review, we briefly discuss the technological advances in Raman and Fourier transform infrared (FT-IR) instrumentation and their application for the analysis of clinically and industrially relevant microorganisms, biofilms, and biological warfare agents. In addition, we summarize the current trends and future prospects of integrating Raman/SERS-isotopic labeling and cell sorting technologies in parallel, to link genotype-to-phenotype in order to define community function of unculturable microbial cells in mixed microbial communities which possess admirable traits such as detoxification of pollutants and recycling of essential metals. -

Spectral Artefacts Induced by Moving Targets in Live Hyperspectral Stimulated Raman Spectroscopy: the Case of Lipid Droplets in Yeast Cells
Clinical Spectroscopy 3 (2021) 100014 Contents lists available at ScienceDirect Clinical Spectroscopy journal homepage: www.sciencedirect.com/journal/clinical-spectroscopy Spectral artefacts induced by moving targets in live hyperspectral stimulated Raman spectroscopy: The case of lipid droplets in yeast cells Cassio Lima a, Chrispian W. Theron a, Howbeer Muhamadali a, Douglas B. Kell a,b, Royston Goodacre a,* a Centre for Metabolomics Research, Department of Biochemistry and Systems Biology, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool, L69 7ZB, United Kingdom b Novo Nordisk Foundation Centre for Biosustainability, Technical University of Denmark, Building 220, Kemitorvet, 2800, Kgs Lyngby, Denmark ARTICLE INFO ABSTRACT Keywords: In this study, we used stimulated Raman spectroscopy (SRS) microscopy to collect Raman signatures from live Yeast Saccharomyces cerevisiae cells in the spectral range 2804 3060 cm 1 and 830 2000 cm 1 with a spectral res Hyperspectral SRS olution of 8 cm 1. To effect this, we tuned the pump beam to several distinct wavelengths and thus acquired a Fixatives series of chemical maps in order to reconstruct SRS spectra based on the intensity of the pixels, an approach also Live cell imaging referred as hyperspectral SRS (hsSRS). One of the advantages of hsSRS over spontaneous Raman is that it is not Raman overtly plagued by fluorescence and so fluorescent samples like yeast can be analysed. We show however that Raman signatures acquired by this approach may be subject to spectral artefacts that manifest as drops in in tensity of Raman signal due to the movement of lipid droplets (LDs) within the yeast cells. -

Cucumis Melo, Cucurbitaceae) Biodiversity
H OH metabolites OH Article Comparative Metabolomics and Molecular Phylogenetics of Melon (Cucumis melo, Cucurbitaceae) Biodiversity Annick Moing 1 , J. William Allwood 2 , Asaph Aharoni 3, John Baker 4, Michael H. Beale 4, Shifra Ben-Dor 3 , Benoît Biais 1, Federico Brigante 5,6,7, Yosef Burger 8, Catherine Deborde 1 , Alexander Erban 5, Adi Faigenboim 8, Amit Gur 9, Royston Goodacre 10 , Thomas H. Hansen 11, Daniel Jacob 1, Nurit Katzir 9, Joachim Kopka 5, Efraim Lewinsohn 9, Mickael Maucourt 1 , Sagit Meir 3 , Sonia Miller 4, Roland Mumm 12, Elad Oren 9, Harry S. Paris 9, Ilana Rogachev 3, Dominique Rolin 1, Uzi Saar 9, Jan K. Schjoerring 11, Yaakov Tadmor 9, Galil Tzuri 9, 12 4 8 12,13, Ric C.H. de Vos , Jane L. Ward , Elena Yeselson , Robert D. Hall y and 8, , Arthur A. Schaffer * y 1 INRAE, Univ. Bordeaux, UMR1332 Fruit Biology and Pathology, Bordeaux Metabolome Facility MetaboHUB, Centre INRAE de Nouvelle Aquitaine - Bordeaux, 33140 Villenave d’Ornon, France; [email protected] (A.M.); [email protected] (B.B.); [email protected] (C.D.); [email protected] (D.J.); [email protected] (M.M.); [email protected] (D.R.) 2 The James Hutton Institute, Environmental & Biochemical Sciences, Invergowrie, Dundee, DD2 5DA Scotland, UK; [email protected] 3 Department of Plant and Environmental Sciences, Weizmann Institute of Science, Rehovot 7610001, Israel; [email protected] (A.A.); [email protected] (S.M.); [email protected] (I.R.); [email protected] (S.B.-D.) 4 Rothamsted Research, Harpenden, Herts AL5 2JQ, UK; john.baker@pfizer.com (J.B.); [email protected] (M.H.B.); [email protected] (S.M.); [email protected] (J.L.W.) 5 Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm 14476, Germany; [email protected] (F.B.); [email protected] (A.E.); [email protected] (J.K.) 6 Universidad Nacional de Córdoba, Facultad de Ciencias Químicas, Dto. -

Portable Through Bottle SORS for the Authentication of Extra Virgin Olive Oil
applied sciences Article Portable through Bottle SORS for the Authentication of Extra Virgin Olive Oil Mehrvash Varnasseri 1, Howbeer Muhamadali 1, Yun Xu 1, Paul I. C. Richardson 1, Nick Byrd 2, David I. Ellis 3 , Pavel Matousek 4 and Royston Goodacre 1,* 1 Department of Biochemistry and Systems Biology, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, BioSciences Building, Crown St., Liverpool L69 7ZB, UK; [email protected] (M.V.); [email protected] (H.M.); [email protected] (Y.X.); [email protected] (P.I.C.R.) 2 Campden BRI Group, Station Road, Chipping Campden GL55 6LD, UK; [email protected] 3 School of Chemistry, Manchester Institute of Biotechnology, University of Manchester, Manchester M1 7DN, UK; [email protected] 4 Central Laser Facility, Research Complex at Harwell, STFC Rutherford Appleton Laboratory, Harwell Oxford OX11 0QX, UK; [email protected] * Correspondence: [email protected] Abstract: The authenticity of olive oil has been a significant long-term challenge. Extra virgin olive oil (EVOO) is the most desirable of these products and commands a high price, thus unscrupulous individuals often alter its quality by adulteration with a lower grade oil. Most analytical methods employed for the detection of food adulteration require sample collection and transportation to a central laboratory for analysis. We explore the use of portable conventional Raman and spatially-offset Citation: Varnasseri, M.; Raman spectroscopy (SORS) technologies as non-destructive approaches to assess the adulteration Muhamadali, H.; Xu, Y.; Richardson, status of EVOO quantitatively and for SORS directly through the original container, which means P.I.C.; Byrd, N.; Ellis, D.I.; Matousek, that after analysis the bottle is intact and the oil would still be fit for use. -

Mass Spectrometry Als
Vol. 15 No. 3 July/September 2019 SPECTROSCOPY asia The essential magazine for spectroscopists in the Asia/Pacific region Global Hg pollution from artisanal gold mining Soil organic matter using vis-NIR spectroscopy Are omics the death of Good Sampling Practice? Vol. 15 No. 3 July/September 2019 ISSN 1743-5277 SPECTROSCOPY asia The essential magazine for spectroscopists in the Asia/Pacific region Global Hg pollution from artisanal gold mining Soil organic matter using vis-NIR spectroscopy CONTENTS Are omics the death of Good Sampling Practice? 3 Editorial 4 News Karl Norris: Father of NIR spectroscopy; First MS images recorded using MAIV; Terahertz QCL without cryogenic cooling Soil organic matter can be determined Reducing global mercury pollution using visible-near infrared spectroscopy and 6 machine learning, providing a green method- from small-scale artisanal gold mining ology for the analysis. Find out more in the article starting on page 11. Peter W.U. Appel and Kim H. Esbensen 11 Determination of soil organic Publisher matter using visible-near infrared Ian Michael ([email protected]) spectroscopy and machine learning Felipe Bachion de Santana, Sandro Keiichi Otani, André Marcelo de Souza and Advertising Sales Ronei Jesus Poppi UK and Ireland Ian Michael Spectroscopy Asia, 6 Charlton Mill, Charlton, Chichester, West Sussex PO18 0HY, 15 Tony Davies Column: Are omics United Kingdom. Tel: +44-1243-811334, Fax: +44-1243-811711, the death of Good Sampling Practice? E-mail: [email protected] Antony N. Davies and Roy Goodacre Americas Joe Tomaszewski John Wiley & Sons Inc. 18 Sampling Column: Passing a Tel: +1-908-514-0776 E: [email protected] milestone: the successful WCSB9 Lili Jiang and Kim H. -

2020-10 Bbcon Complete Programme RGB 28Oct
Welcome to the: Digital Breath Biopsy Conference 10th - 11th NOVEMBER 2020 #BBcon20 Programme (subject to change) DAY 1 Tuesday 10th November 2020 TIME (GMT) EVENT 9:45-10:00 Welcome to the Breath Biopsy Conference Session 1: Applications of VOCs Alan Griths, LECO The timeline of breath analysis - a brief history of when, why and how Stephen Fowler, University of Manchester Breath analysis - not (only) for diagnosis Agi Smolinska, Maastricht University 10:00-12:15 Exhaled breath as early predictiveEVENT signatures of asthma in children James Covington, University of Warwick The diagnosis and monitoring of inflammatory bowel disease by the analysis of breath and other human waste Anthony Hobson, The Functional Gut Clinic Digestion, fermentation and function – Using breath analysis to assess digestive health 12:15-12:45 Break and Networking Session 2: Breath Biomarkers for Liver Disease Giuseppe Ferrandino, Owlstone Medical Breath Biopsy to discover novel exogenous volatile organic compound (EVOC) biomarkers for chronic liver disease 12:45-14:15 John Plevris, University of Edinburgh Volatomic analysis has the potential to non-invasively and accurately stratify patients with non-alcoholic fatty liver disease Scott Friedman, Icahn School of Medicine at Mount Sinai Diagnostic challenges in non-alcoholic fatty liver disease (NAFLD) 14:15-14:45 Break and Networking Poster Session & Keynote Talks Poster presentations 5 minute presentations of selected conference posters 14:45-17:15 Terence Risby, Johns Hopkins Bloomberg School of Public Health -

Raman Spectroscopy As a Diagnostic Tool 0003-2654(2013)138:14;1-X Analyst
Volume 138 | Number 14 | 2013 138 | Number Volume Analyst www.rsc.org/analyst Volume 138 | Number 14 | 21 July 2013 | Pages 3847–4204 Analyst Themed issue: Optical Diagnosis Showcasing Raman detection of early stages of As featured in: malaria infection from the Biophotonics Laboratory of Nicholas Smith at the Immunology Frontier Research Center, Osaka University, Japan. Title: Raman spectroscopic analysis of malaria disease progression via blood and plasma samples We show that Raman spectroscopy can discriminate between resonant heme and bio-crystallized hemozoin in the blood of malaria-infected patients. Both are present in blood and plasma following infection, and by multivariate analysis, their discrimination within blood samples allows early, quantitative and See Nicholas I. Smith et al., potentially automated detection of malaria, and its eff ects on the Analyst, 2013, 138, 3927. immune system. Pages 3847–4204 Pages Themed issue: Optical Diagnosis www.rsc.org/analyst ISSN 0003-2654 Registered Charity Number 207890 CRITICAL REVIEW David I. Ellis et al. Illuminating disease and enlightening biomedicine: Raman spectroscopy as a diagnostic tool 0003-2654(2013)138:14;1-X Analyst CRITICAL REVIEW Illuminating disease and enlightening biomedicine: Raman spectroscopy as a diagnostic tool Cite this: Analyst, 2013, 138, 3871 David I. Ellis,*a David P. Cowcher,a Lorna Ashton,a Steve O'Hagana and Royston Goodacreab The discovery of the Raman effect in 1928 not only aided fundamental understanding about the quantum nature of light and matter but also opened up a completely novel area of optics and spectroscopic research that is accelerating at a greater rate during the last decade than at any time since its inception. -
Metabolomics
@RoyGoodacre @LivUniCMR www.biospec.net @Metabolomics Metabolomics by numbers: lessons from large-scale phenotyping Roy Goodacre and friends From metabolites to metabolomics Human Metabolism Metabolite Metabolomics intermediate defined as the metabolic of metabolism complement (metabolite pool) of a cell or tissue type under a given set of conditions “Traditional” linear view of a metabolic pathway A “scale-free” metabolic network Metabolomics & biological systems Volatilome metabolites Endo-metabolome Biopsy metabolomics Sputum 1y cell culture S cells footprint Biofluids (exo-metabolome) pathways networks proteins/mRNA Integrate: SNP / genotype -----> system understanding www.husermet.org Funded by and in collaboration with: Department of Trade and Industry GC-MS + RPLC-MS METABOLIC METABOLIC PATHWAYS PATHWAYS Glycolysis, TCA cycle Lipid and fatty acid Pentose Phosphate metabolism Amino acid metabolism Secondary metabolite synthesis Gluconeogenesis Metabolism of co- Urea cycle factors and vitamins Inositol metabolism Metabolism of Xenobiotics Carbohydrate metabolism PROVIDES GOOD METABOLITE COVERAGE IN COMPLEMENTARY PATHWAYS LESSON I Mass Spectrometers & Chromatography DRIFT QCs allow signal assessment QC 1 QC 2 Real: QC 3 QC 4 60 serum (healthy). QC 5 QC 6 QA: QC 7 sigma serum. QC 8 QC 9 Spike1: QC 10 blank glutaric acid, citric column test solution acid, alanine, glycine, sample 1 sample 2 leucine, sample 3 phenylalanine, and sample 4 sample 5 tryptophan. QC 11 -1 [each at 0.16 mg mL ] sample 6 sample 7 Spike 2: sample 8 caffeine & nicotine. sample 9 sample 10 [each at 0.16 mg mL-1] QC 12 sample 11 Begley P. et al. (2009) Analytical Chemistry 71, 7038-7046 QCs allow signal correction Instrument annual maintenance Sample QC QC 1 Before: QC 2 QC 3 QC 4 QC 5 QC 6 QC 7 QC 8 QC 9 QC 10 blank After: column test solution sample 1 sample 2 sample 3 sample 4 sample 5 QC 11 sample 6 sample 7 sample 8 sample 9 LOESS: low-order nonlinear locally estimated smoothing function sample 10 QC 12 sample 11 Dunn W. -
Inside Raman UK Seminar 8Th and 9Th April 2014, Manchester Conference Centre
www.renishaw.com/ir2014 Inside Raman UK seminar 8th and 9th April 2014, Manchester Conference Centre Previous attendees said… “Very interesting talks that enlighten wider use of Raman in science” “Diverse program of speakers” “Really useful to see how current users utilise the abilities of their Renishaw instruments” “Opportunity to network and find out about applications of Raman” “Helpful to talk to other users and the Renishaw applications experts” Inside Raman UK seminar Discover more with inVia Share your experience and discover new ideas as we bring together Raman users and experts from a diverse range of applications. Featuring presentations from prominent scientists on their research involving Raman spectroscopy, Inside Raman will appeal to anyone interested in learning about—or extending their understanding of—this technique. Great networking There will be plenty of opportunities for you to speak with colleagues from both academia and industry. You will also see demonstrations of Renishaw’s latest technology, capabilities and software. Programme The seminar takes place over two days and caters for people with all levels of Raman knowledge and experience, from students to postdocs and researchers. Inside Raman is a fantastic learning and networking opportunity. It includes: • a variety of talks from prominent scientists • demonstrations by Renishaw’s applications team • a contributed poster session (on day one) At the end of day one, all delegates are invited to join us for a relaxed dinner, at the Manchester Conference Centre. 8th and 9th April 2014 Manchester Conference Centre Invited speakers Inside Raman features presentations from prominent scientists. The following speakers have been confirmed and more will be added.