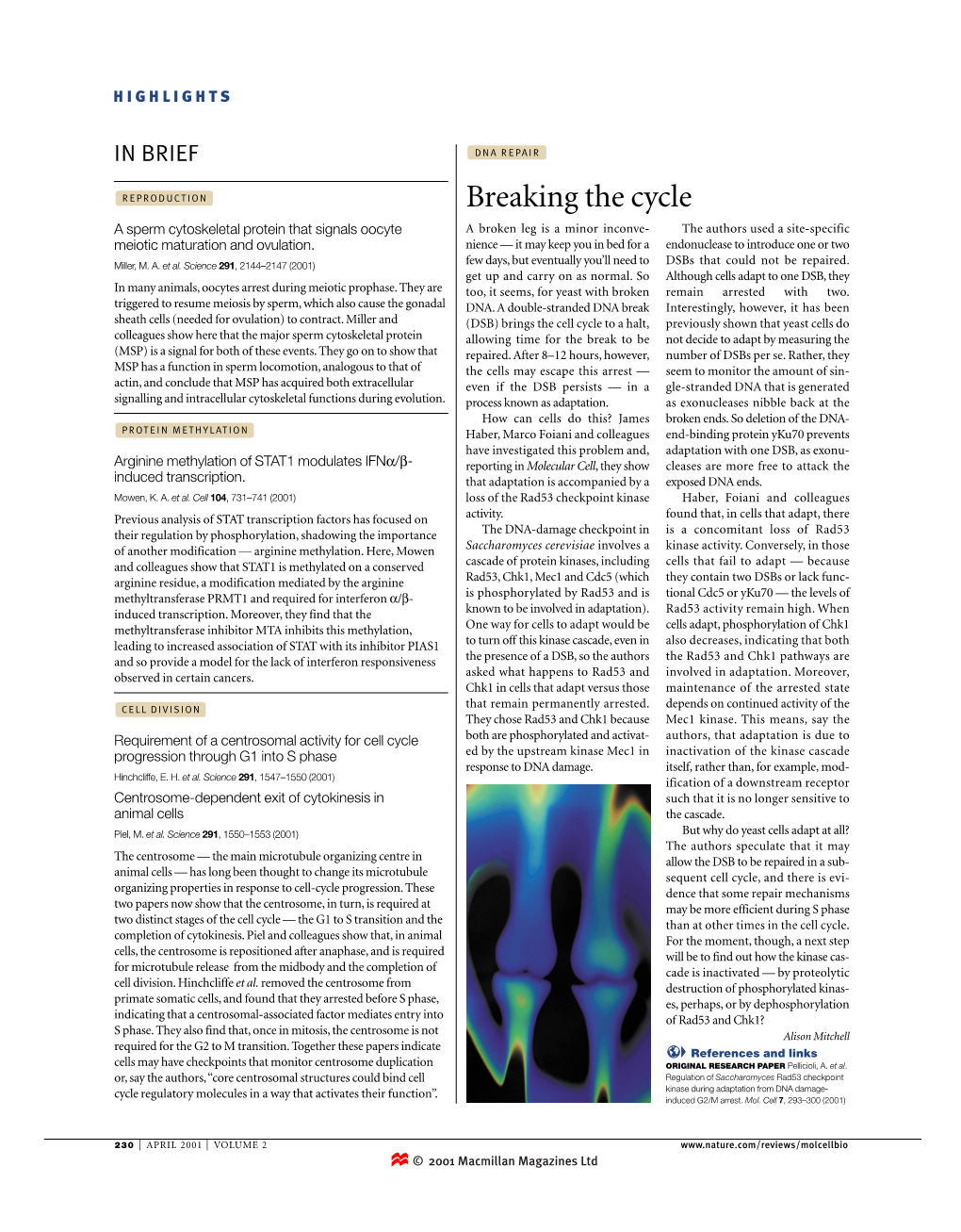
Breaking the Cycle a Sperm Cytoskeletal Protein That Signals Oocyte a Broken Leg Is a Minor Inconve- the Authors Used a Site-Specific Meiotic Maturation and Ovulation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

3 Saccharomyces Cell Structure
Genetic Techniques for Biological Research Corinne A. Michels Copyright q 2002 John Wiley & Sons, Ltd ISBNs: 0-471-89921-6 (Hardback); 0-470-84662-3 (Electronic) 3 Saccharomyces Cell Structure Saccharomycescerevisiue is aeukaryote and as such containsthe subcellular organelles commonlyfound in eukaryotes.The structure and function of these organelles is fundamentally the same as it is in other eukaryotes with less versatile systems for genetic analysis, and for this reason Saccharomyces is the organism of choice for many cell biologists. For a truly in-depth review of Saccharomyces cell structure and function, the reader is referred to The Molecular and Cellular Biology of theYeast Saccharomyces: Vol. 3: CellCycle and Cell Biology (Broach etal., 1997). The discussion here will provide a very brief overview of the cell structure and will focus on certain unique features of Saccharomyces cell structure in order to facilitate reading of the literature. CELL SHAPE AND GROWTH PATTERNS Underusual culture conditions, Saccharomyces is ellipsoidaUovoid in shapeand approximately 5-10 pm long by 3-7 pm wide. This is referred to as the yeast form. Figure 3.1 shows a scanning electron micrograph (SEM) of a cell in the yeast form. Cell division is by budding; that is, a smaller ovoid daughter cell formsas a projection from the surface of the mother cell. Haploid cells are generally about one-half the volume of diploid cells. The characteristic shape is maintained by a rigid cell wall that completely surrounds the plasma membrane of Saccharomyces. Changes in this shape involve remodeling of the cell wall and occur during budding, mating,and pseudohyphal differentiation. -

The Ultrastructure of Spermatozoa and Spermiogenesis in Pyramidellid Gastropods, and Its Systematic Importance John M
HELGOLANDER MEERESUNTERSUCHUNGEN Helgol~inder Meeresunters. 42,303-318 (1988) The ultrastructure of spermatozoa and spermiogenesis in pyramidellid gastropods, and its systematic importance John M. Healy School of Biological Sciences (Zoology, A08), University of Sydney; 2006, New South Wales, Australia ABSTRACT: Ultrastructural observations on spermiogenesis and spermatozoa of selected pyramidellid gastropods (species of Turbonilla, ~gulina, Cingufina and Hinemoa) are presented. During spermatid development, the condensing nucleus becomes initially anterio-posteriorly com- pressed or sometimes cup-shaped. Concurrently, the acrosomal complex attaches to an electron- dense layer at the presumptive anterior pole of the nucleus, while at the opposite (posterior) pole of the nucleus a shallow invagination is formed to accommodate the centriolar derivative. Midpiece formation begins soon after these events have taken place, and involves the following processes: (1) the wrapping of individual mitochondria around the axoneme/coarse fibre complex; (2) later internal metamorphosis resulting in replacement of cristae by paracrystalline layers which envelope the matrix material; and (3) formation of a glycogen-filled helix within the mitochondrial derivative (via a secondary wrapping of mitochondria). Advanced stages of nuclear condensation {elongation, transformation of fibres into lamellae, subsequent compaction) and midpiece formation proceed within a microtubular sheath ('manchette'). Pyramidellid spermatozoa consist of an acrosomal complex (round -

Formation and Structure of Filamentous Systems for Insect Flight and Mitotic Movements Melvyn Dennis Goode Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1967 Formation and structure of filamentous systems for insect flight and mitotic movements Melvyn Dennis Goode Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Genetics Commons Recommended Citation Goode, Melvyn Dennis, "Formation and structure of filamentous systems for insect flight and mitotic movements " (1967). Retrospective Theses and Dissertations. 3391. https://lib.dr.iastate.edu/rtd/3391 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. FORMATION AND STRUCTURE OF FILAMENTOUS SYSTEMS FOR INSECT FLIGHT AND MITOTIC MOVEMENTS by Melvyn Dennis Goode A Dissertation Submitted to the Graduate Faculty in Partial Fulfillment of The Requirements for the Degree of DOCTOR OF PHILOSOPHY Major Subject: Cell Biology Approved: Signature was redacted for privacy. In Charge oi Major Work Signature was redacted for privacy. Chairman Advisory Committee Cell Biology Program Signature was redacted for privacy. Signature was redacted for privacy. Iowa State University Of Science and Technology Ames, Iowa: 1967 ii TABLE OF CONTENTS Page I. INTRODUCTION 1 PART ONE. THE MITOTIC APPARATUS OF A GIANT AMEBA 5 II. THE STRUCTURE AND PROPERTIES OF THE MITOTIC APPARATUS 6 A. Introduction .6 1. Early studies of mitosis 6 2. The mitotic spindle in living cells 7 3. -

Ochromonas Mitochondria Contain a Specific Chloroplast Protein
Proc. Nati. Acad. Sci. USA Vol. 82, pp. 1456-1459, March 1985 Cell Biology Ochromonas mitochondria contain a specific chloroplast protein (small subunit of ribulose-1,5-blsphosphate carboxylase/immunoelectron microscopy/chrysophycean alga/promiscuous DNA) GINETTE LACOSTE-ROYAL AND SARAH P. GIBBS Department of Biology, McGill University, Montreal, Quebec H3A iBi, Canada Communicated by Lynn Margulis, November 1, 1984 ABSTRACT Antibody raised against the small subunit of at 200C in Beijerinck's medium (15) at a light intensity of ribulose-1,5-bisphosphate carboxylase [3-phospho-D-glycerate 4300 lux. carboxy-lyase (dimerizing), EC 4.1.1.39] of Chlamydomonas Fixation and Embedding. Cells were fixed in 1% (vol/vol) reinhardtii labeled the mitochondria as well as the chloroplast glutaraldehyde in 0.1 M phosphate buffer for 90 min at 40C, of the chrysophyte alga Ochromonas danica in sections pre- rinsed in buffer and blocked in 2% (wt/vol) agar. The agar pared for immunoelectron microscopy by the protein A-gold blocks were dehydrated in 25% (vol/vol) ethanol at -50C, technique. The same antibody labeled the chloroplast but not then in 50%, 75%, and 95% ethanol at -18'C. Embedding the mitochondria of C. reinhardti. A quantitative study of la- was carried out at -18'C in Lowicryl K4M (16) according to beling in dark-grown, greening (32 hr light), and mature green the following schedule: 95% ethanol/resin, 1:1 (vol/vol), cells of 0. danica revealed that anti-small-subunit staining in overnight; 95% ethanol/resin, 1:2 (vol/vol), 2 times for 2 hr the mitochondria increased progressively in the light as it does each; pure resin, 2 hr and then overnight. -

The Role of Endoplasmic Reticulum in the Repair of Amoeba Nuclear Envelopes Damaged Microsurgically
J. Cell Sci. 14, 421-437 ('974) 421 Printed in Great Britain THE ROLE OF ENDOPLASMIC RETICULUM IN THE REPAIR OF AMOEBA NUCLEAR ENVELOPES DAMAGED MICROSURGICALLY C. J. FLICKINGER Department of Anatomy, School of Medicine, University of Virginia, CharlottesvilU, Virginia 22901, U.S.A. SUMMARY The nuclear envelopes of amoebae were damaged microsurgically, and the fate of the lesions was studied with the electron microscope. Amoebae were placed on the surface of an agar- coated slide. Using a glass probe, the nucleus was pushed from an amoeba, damaged with a chopping motion of the probe, and reinserted into the amoeba. Cells were prepared for electron microscopy at intervals of between 10 min and 4 days after the manipulation. Nuclear envelopes studied between 10 min and 1 h after the injury displayed extensive damage, includ- ing numerous holes in the nuclear membranes. Beginning 15 min after the manipulation, pieces of rough endoplasmic reticulum intruded into the holes in the nuclear membranes. These pieces of rough endoplasmic reticulum subsequently appeared to become connected to the nuclear membranes at the margins of the holes. By 1 day following the injury, many cells had died, but the nuclear membranes were intact in those cells that survived. The elaborate fibrous lamina or honeycomb layer characteristic of the amoeba nuclear envelope was resistant to early changes after the manipulation. Patches of disorganization of the fibrous lamina were present 5 h to 1 day after injury, but the altered parts showed evidence of progress toward a return to normal configuration by 4 days after the injury. It is proposed that the rough endoplasmic reticulum participates in the repair of injury to the nuclear membranes. -

Flagella Apparatus
MOTILE CELL Characters and Character States Location code / cell type ZO, zoospore Flagella apparatus Kinetosome Electron-opaque material in core in kinetosome 0, absent (everything else); 1, present (Kappamyces). Electron-opaque material in axoneme core and between axoneme and flagellar membrane 0, absent; 1, present (many Chytridiales). Kinetosome characters Scalloped ring within kinetosome, extensions of the A, B, or C microtubule Flagellum coating 0, absent; 1, present (Polyphagus euglenae). Number of flagella 0, one 1, multiple Kinetosome-associated structures (KAS) Kinetosome support 0, absent 1, kinetosome props; 2, broken kinetosome props (Olpidium radicale); 3, saddle-like structure surrounding kinetosome (Neocallimastigales) Kinetosome-associated plates 0, absent; 1, present. Kinetosome-associated spur 0, absent; 1, present. Kinetosome-associated shield 0, absent; 1, present. Kinetosome-associated veil 0, absent; 1, present. Anteriorly oriented kinetosome microtubule organizing center (MTOC) (Spizellomycetales) 0, absent; 1, simple solid, not laminate, only one part (Spizellomyces); 2, stacked plates with radiating anteriorly oriented microtubules (Powellomyces) 3, stacked plates with laterally anteriorly oriented microtubules (Gaerteromyces) 4, compound, two in stacked arrangement (Kochiomyces) 5, tripartitalcar rod with 3 lobes in cross-section Primary microtubule roots 0, absent; 1, 2-5 parallel microtubules are stacked with space between them, no connectors (Rhyzophydium); 2, approx. 6-7 parallel microtubules chord-like; 3, more than 20 parallel microtubules, bundled and have connectors/linkers between them (Nowakowskiella); 4, microtubules posterior to anterior, parallel to kinetosome triplets (Batracomyces – not separated like Rhysophydium). Striated rhizoplast 0, absent; 1, present. Flagellar rootlet absolute configuration 0, absent; 1, 20 degrees left; 2, 10 degrees right; 3, 50 degrees left. -

Establishment of a Fungal Model System for the Study of Ciliation
ESTABLISHMENT OF A FUNGAL MODEL SYSTEM FOR THE STUDY OF CILIATION Linnea Tracy June 2015 ESTABLISHMENT OF A FUNGAL MODEL SYSTEM FOR THE STUDY OF CILIATION An Honors Thesis Submitted to the Department of Biology in partial fulfillment of the Honors Program STANFORD UNIVERSITY by Linnea Tracy June 2015 2 Acknowledgements A simple thank-you seems inadequate for all those who have offered their time, expertise, support, and supplies towards my project and education that is culminating in this thesis. Nevertheless, thank you to Tim Stearns, whose kindness, brilliance, and natural knack for teaching was an inspiration to me as a student, was motivation to me as a researcher, and was a great honor to get to know and work closely with over the last two years. Thank you for your time and dedication devoted to my, my peers’, and the world’s education. A special thank you to Erin Turk, who took me under her wing, learning about chytrids in order to teach and assist me, all the while completing her own graduate dissertation. You are one of the most motivated, prepared, and lovely people I have met at Stanford. Thank you for your guidance, mentorship, and always responding to my text messages. Thank you to the members of the Stearns lab, who guided me from learning the basics of the laboratory through experimental design and science writing. It has been, and continues to be, a great pleasure to have a lab family that is so intelligent and kind. I would be remiss to not also thank my parents and friends, who have loyally allowed me to discuss cilia and worry over my experiments with them, sometimes at the expense of our social life. -

Direct Continuities Between Cisternae at Different Levels of the Golgi Complex in Glucose-Stimulated Mouse Islet Beta Cells
Direct continuities between cisternae at different levels of the Golgi complex in glucose-stimulated mouse islet beta cells Brad J. Marsh*†, Niels Volkmann‡, J. Richard McIntosh*, and Kathryn E. Howell§ *Boulder Laboratory for 3D Electron Microscopy of Cells, Department of Molecular, Cellular, and Developmental Biology, University of Colorado, Boulder, CO 80309; ‡The Burnham Institute, 10901 North Torrey Pines Road, La Jolla, CA 92037; and §Department of Cellular and Structural Biology, University of Colorado Health Sciences Center, University of Colorado School of Medicine, Denver, CO 80262 Communicated by Pamela J. Bjorkman, California Institute of Technology, Pasadena, CA, February 23, 2004 (received for review January 10, 2004) Direct continuity between the membranes of cisternae in the Golgi Thus, the presence or absence of cisternal continuity, previously complex in mammalian cells rarely has been observed; when seen, thought to reflect differences in experimental technique, may in its documentation has been equivocal. Here we have used dual- fact represent different states adopted by the cells’ secretory axis electron microscope tomography to examine the architecture machinery as a function of different levels of protein synthesis of the Golgi in three dimensions at Ϸ6-nm resolution in rapidly and transport. frozen, freeze-substituted murine cells that make and secrete insulin in response to glucose challenge. Our data show three types Materials and Methods of direct connections between Golgi cisternae that are normally Islet Isolation and Culture. Intact islets of Langerhans obtained distinct from one another. These connections all ‘‘bypass’’ inter- from the pancreata of freshly killed adult, female, BALB͞c mice, ceding cisternae. We propose that when pancreatic beta cells are were used to ensure that neither cellular polarity nor extracel- stimulated to synthesize and secrete insulin rapidly in vivo, such lular matrices were disrupted. -

Microtubular Organization in Elongating Myogenic Cells
MICROTUBULAR ORGANIZATION IN ELONGATING MYOGENIC CELLS ROBERT H. WARREN From the Department of Biological Structure, University of Miami School of Medicine, Miami, Florida 33152 ABSTRACT Microtubule organization has been studied in serially sectioned myogenic cells in the tail muscle regeneration blastema of Rana pipiens tadpoles. In mesenchymal cells and in some premyoblasts, microtubules radiate from centriolar satellites in a cell center, while in more mature myoblasts and myotubes the centrioles no longer appear to serve as organizing centers for microtubules. In all elongate, fusiform myogenic cells, the microtubules are predominately oriented in the lon- gitudinal axis of the cell. Counts of microtubules in transverse sections spaced at regular intervals along the cells show that the absolute number of microtubules is greatest in the thickened midregions of the cells and decreases relatively smoothly toward the tapered ends of the cells. Close paraxial association of micro- tubules (within 40 nm surface-to-surface) occurs along the entire lengths of cells but appears with greatest frequency in their tapered ends. In two myoblasts, serial sections were used to trace all microtubules in 8-~m long segments of the cells located about midway between the nucleus and one end of the cell. Since tracings show that as many as 50% of the microtubules terminate within the 8- /zm long segment, it seems unlikely that any microtubules extend the entire length of the cell. It is proposed that lateral interactions between paraxial micro- tubules stabilize the overall microtubular apparatus and contribute to mainte- nance of the bipolar form of the cells. A three-dimensional model of the complete microtubular array in one of the 8-urn long segments of a myoblast has been con- structed. -

Contacts Between the Endoplasmic Reticulum and Other Membranes In
Contacts between the endoplasmic reticulum and other PNAS PLUS membranes in neurons Yumei Wua,b,c,d, Christina Whiteusa,b,c,d, C. Shan Xue, Kenneth J. Hayworthe, Richard J. Weinbergf, Harald F. Hesse, and Pietro De Camillia,b,c,d,g,1 aDepartment of Neuroscience, Yale University School of Medicine, New Haven, CT 06510; bDepartment of Cell Biology, Yale University School of Medicine, New Haven, CT 06510; cHoward Hughes Medical Institute, Yale University School of Medicine, New Haven, CT 06510; dProgram in Cellular Neuroscience, Neurodegeneration, and Repair, Yale University School of Medicine, New Haven, CT 06510; eJanelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA 20147; fDepartment of Cell and Developmental Biology, Neuroscience Center, University of North Carolina, Chapel Hill, NC 27599; and gKavli Institute for Neurosciences, Yale University School of Medicine, New Haven, CT 06510 Contributed by Pietro De Camilli, May 5, 2017 (sent for review January 23, 2017; reviewed by Kristen M. Harris and Gia Voeltz) Close appositions between the membrane of the endoplasmic variety of functions for these contacts (7–12). One major reticulum (ER) and other intracellular membranes have important function is exchange of lipids between bilayers via lipid-transfer functions in cell physiology. These include lipid homeostasis, proteins (11, 12, 21–24). The occurrence of this mode of lipid + regulation of Ca2 dynamics, and control of organelle biogenesis transfer between membranes, which is independent of vesicular and dynamics. Although these membrane contacts have previ- transport and membrane fusion, was first described in the late ously been observed in neurons, their distribution and abundance 1960s (25). -

Mapping the Distribution of Golgi Enzymes Involved in the Construction of Complex Oligosaccharides
Journal of Cell Science 108, 1617-1627 (1995) 1617 Printed in Great Britain © The Company of Biologists Limited 1995 Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides Catherine Rabouille, Norman Hui, Felicia Hunte, Regina Kieckbusch, Eric G. Berger*, Graham Warren and Tommy Nilsson† Cell Biology Laboratory, Imperial Cancer Research Fund, 44 Lincoln’s Inn Fields, London, WC2A 3PX, UK *Present address: Physiologisches Institut, Universität Zürich, Switzerland †Author for correspondence SUMMARY The distribution of β1,2 N-acetylglucosaminyltransferase I tion but they occupied the trans cisterna and the trans- (NAGT I), α1,3-1,6 mannosidase II (Mann II), β1,4 galac- Golgi network (TGN). These results generalise our earlier tosyltransferase (GalT), α2,6 sialyltransferase (SialylT) observations on the overlapping distribution of Golgi was determined by immuno-labelling of cryo-sections from enzymes and show that each of the trans compartments of HeLa cell lines. Antibody labelling in the HeLa cell line was the Golgi apparatus in HeLa cells contains unique mixtures made possible by stable expression of epitope-tagged forms of those Golgi enzymes involved in the construction of of these proteins or forms from species to which specific complex, N-linked oligosaccharides. antibodies were available. NAGT I and Mann II had the same distribution occupying the medial and trans cisternae Key words: Golgi apparatus, trans-Golgi network, of the stack. GalT and SialylT also had the same distribu- glycosyltransferase, mannosidase, TGN38 INTRODUCTION two adjacent cisternae was taken as evidence of cisternal dupli- cation. This interpretation was supported by the observation The construction of complex, bi-antennary, N-linked oligosac- that the number of cisternae in the Golgi stack can vary widely charides involves the sequential action of enzymes located in from tissue to tissue and from organism to organism (see different parts of the Golgi apparatus (for reviews, see Kornfeld Fawcett, 1981). -

3-D Ultrastructure of O. Tauri: Electron
III-1 3-D Ultrastructure of O. tauri: Electron Cryotomography of an Entire Eukaryotic Cell Gregory P. Henderson, Lu Gan, and Grant J. Jensen* Division of Biology, California Institute of Technology, Pasadena, CA Running Title: 3-D Ultrastructure of Ostreococcus tauri * Corresponding Author. Mail address: Caltech Division of Biology, 1200 E. California Blvd., Pasadena, CA 91125. Phone: (626) 395-5730. Fax: (626) 395-5730. E-mail: [email protected]. III-2 Abstract The hallmark of eukaryotic cells is their segregation of key biological functions into discrete, membrane-bound organelles. Creating accurate models of their ultrastructural complexity has been difficult in part because of the limited resolution of light microscopy and the artifact-prone nature of conventional electron microscopy. Here we explored the potential of the emerging technology electron cryotomography to produce three-dimensional images of an entire eukaryotic cell in a near-native state. Ostreococcus tauri was chosen as the specimen because as a unicellular picoplankton with just one copy of each organelle, it is the smallest known eukaryote and was therefore likely to yield the highest resolution images. Whole cells were imaged at various stages of the cell cycle, yielding 3-D reconstructions of complete chloroplasts, mitochondria, endoplasmic reticula, Golgi bodies, peroxisomes, microtubules, and putative ribosome distributions in-situ. Surprisingly, the nucleus was seen to open long before mitosis, and while one microtubule (or two in some predivisional cells) were consistently present, no mitotic spindle was ever observed, prompting speculation that a single microtubule might be sufficient to segregate multiple chromosomes. Abbreviations 3-D, three-dimensional; ECT, electron cryotomography; ER, endoplasmic reticulum; NE, nuclear envelope; NPC, nuclear pore complex.