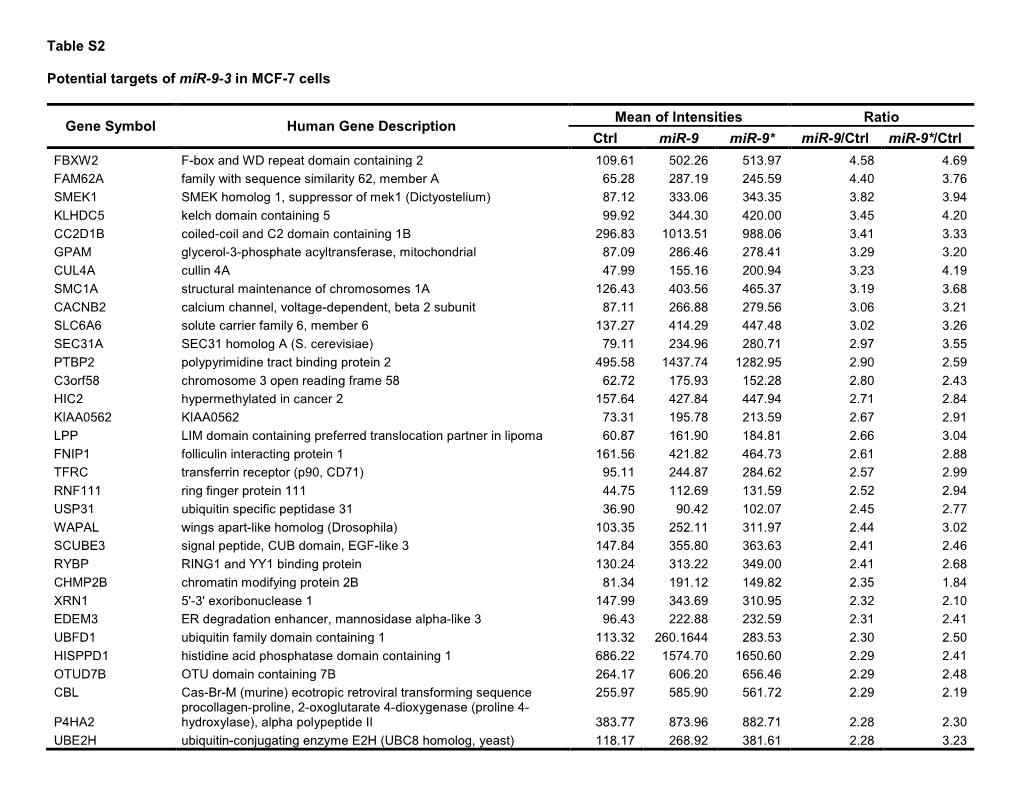
Supplementary Table 2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The HECT Domain Ubiquitin Ligase HUWE1 Targets Unassembled Soluble Proteins for Degradation
OPEN Citation: Cell Discovery (2016) 2, 16040; doi:10.1038/celldisc.2016.40 ARTICLE www.nature.com/celldisc The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation Yue Xu1, D Eric Anderson2, Yihong Ye1 1Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA; 2Advanced Mass Spectrometry Core Facility, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA In eukaryotes, many proteins function in multi-subunit complexes that require proper assembly. To maintain complex stoichiometry, cells use the endoplasmic reticulum-associated degradation system to degrade unassembled membrane subunits, but how unassembled soluble proteins are eliminated is undefined. Here we show that degradation of unassembled soluble proteins (referred to as unassembled soluble protein degradation, USPD) requires the ubiquitin selective chaperone p97, its co-factor nuclear protein localization protein 4 (Npl4), and the proteasome. At the ubiquitin ligase level, the previously identified protein quality control ligase UBR1 (ubiquitin protein ligase E3 component n-recognin 1) and the related enzymes only process a subset of unassembled soluble proteins. We identify the homologous to the E6-AP carboxyl terminus (homologous to the E6-AP carboxyl terminus) domain-containing protein HUWE1 as a ubiquitin ligase for substrates bearing unshielded, hydrophobic segments. We used a stable isotope labeling with amino acids-based proteomic approach to identify endogenous HUWE1 substrates. Interestingly, many HUWE1 substrates form multi-protein com- plexes that function in the nucleus although HUWE1 itself is cytoplasmically localized. Inhibition of nuclear entry enhances HUWE1-mediated ubiquitination and degradation, suggesting that USPD occurs primarily in the cytoplasm. -

A Family of Mammalian E3 Ubiquitin Ligases That Contain the UBR Box Motif and Recognize N-Degrons Takafumi Tasaki,1 Lubbertus C
MOLECULAR AND CELLULAR BIOLOGY, Aug. 2005, p. 7120–7136 Vol. 25, No. 16 0270-7306/05/$08.00ϩ0 doi:10.1128/MCB.25.16.7120–7136.2005 Copyright © 2005, American Society for Microbiology. All Rights Reserved. A Family of Mammalian E3 Ubiquitin Ligases That Contain the UBR Box Motif and Recognize N-Degrons Takafumi Tasaki,1 Lubbertus C. F. Mulder,2 Akihiro Iwamatsu,3 Min Jae Lee,1 Ilia V. Davydov,4† Alexander Varshavsky,4 Mark Muesing,2 and Yong Tae Kwon1* Center for Pharmacogenetics and Department of Pharmaceutical Sciences, School of Pharmacy, University of Pittsburgh, Pittsburgh, Pennsylvania 152611; Aaron Diamond AIDS Research Center, The Rockefeller University, New York, New York 100162; Protein Research Network, Inc., Yokohama, Kanagawa 236-0004, Japan3; and Division of Biology, California Institute of Technology, Pasadena, California 911254 Received 15 March 2005/Returned for modification 27 April 2005/Accepted 13 May 2005 A subset of proteins targeted by the N-end rule pathway bear degradation signals called N-degrons, whose determinants include destabilizing N-terminal residues. Our previous work identified mouse UBR1 and UBR2 as E3 ubiquitin ligases that recognize N-degrons. Such E3s are called N-recognins. We report here that while double-mutant UBR1؊/؊ UBR2؊/؊ mice die as early embryos, the rescued UBR1؊/؊ UBR2؊/؊ fibroblasts still retain the N-end rule pathway, albeit of lower activity than that of wild-type fibroblasts. An affinity assay for proteins that bind to destabilizing N-terminal residues has identified, in addition to UBR1 and UBR2, a huge (570 kDa) mouse protein, termed UBR4, and also the 300-kDa UBR5, a previously characterized mammalian E3 known as EDD/hHYD. -

The Ubiquitin Proteasome System and Its Involvement in Cell Death Pathways
Cell Death and Differentiation (2010) 17, 1–3 & 2010 Macmillan Publishers Limited All rights reserved 1350-9047/10 $32.00 www.nature.com/cdd Editorial The ubiquitin proteasome system and its involvement in cell death pathways F Bernassola1, A Ciechanover2 and G Melino1,3 Cell Death and Differentiation (2010) 17, 1–3; doi:10.1038/cdd.2009.189 Following the awarding of the 2004 Nobel Prize in Chemistry Inactivation of the proteasome following caspase-mediated to Aaron Ciechanover, Avram Hershko, and Irwin A Rose cleavage may disable the proteasome, interfering with its for the discovery of ubiquitin (Ub)-mediated degradation, role in the regulation of key cellular processes and thereby Cell Death and Differentiation has drawn the attention of facilitating induction of apoptosis. The noted recent develop- its readers to the Ub Proteasome System (UPS) and its ments show how understanding of these functions is just involvement in regulating cell death pathways.1–4 The current starting to emerge. For example, why does dIAP1 associate set of reviews is an update on this theme.5–16 with multiple E2s via its RING finger? Does dIAP1 also interact From previous review articles published in Cell Death and with the E3 – the F-box protein Morgue, which is a part of an Differentiation, it was apparent that the UPS has a major SCF E3 complex? Why does dIAP1, which is an E3, have to mechanistic role in regulating cell death via modification and interact with other ligases such as the N-end rule UBR1 and degradation of key regulatory proteins involved in -

Transcriptome Sequencing and Genome-Wide Association Analyses Reveal Lysosomal Function and Actin Cytoskeleton Remodeling in Schizophrenia and Bipolar Disorder
Molecular Psychiatry (2015) 20, 563–572 © 2015 Macmillan Publishers Limited All rights reserved 1359-4184/15 www.nature.com/mp ORIGINAL ARTICLE Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder Z Zhao1,6,JXu2,6, J Chen3,6, S Kim4, M Reimers3, S-A Bacanu3,HYu1, C Liu5, J Sun1, Q Wang1, P Jia1,FXu2, Y Zhang2, KS Kendler3, Z Peng2 and X Chen3 Schizophrenia (SCZ) and bipolar disorder (BPD) are severe mental disorders with high heritability. Clinicians have long noticed the similarities of clinic symptoms between these disorders. In recent years, accumulating evidence indicates some shared genetic liabilities. However, what is shared remains elusive. In this study, we conducted whole transcriptome analysis of post-mortem brain tissues (cingulate cortex) from SCZ, BPD and control subjects, and identified differentially expressed genes in these disorders. We found 105 and 153 genes differentially expressed in SCZ and BPD, respectively. By comparing the t-test scores, we found that many of the genes differentially expressed in SCZ and BPD are concordant in their expression level (q ⩽ 0.01, 53 genes; q ⩽ 0.05, 213 genes; q ⩽ 0.1, 885 genes). Using genome-wide association data from the Psychiatric Genomics Consortium, we found that these differentially and concordantly expressed genes were enriched in association signals for both SCZ (Po10 − 7) and BPD (P = 0.029). To our knowledge, this is the first time that a substantially large number of genes show concordant expression and association for both SCZ and BPD. Pathway analyses of these genes indicated that they are involved in the lysosome, Fc gamma receptor-mediated phagocytosis, regulation of actin cytoskeleton pathways, along with several cancer pathways. -

TRAIP Modulates the IGFBP3/AKT Pathway to Enhance the Invasion
www.nature.com/cddis ARTICLE OPEN TRAIP modulates the IGFBP3/AKT pathway to enhance the invasion and proliferation of osteosarcoma by promoting KANK1 degradation ✉ ✉ Mi Li1,6, Wei Wu2,6, Sisi Deng3, Zengwu Shao2 and Xin Jin 4,5 © The Author(s) 2021 Osteosarcoma is one of the most common primary malignancies in bones and is characterized by high metastatic rates. Circulating tumor cells (CTCs) derived from solid tumors can give rise to metastatic lesions, increasing the risk of death in patients with cancer. Here, we used bioinformatics tools to compare the gene expression between CTCs and metastatic lesions in osteosarcoma to identify novel molecular mechanisms underlying osteosarcoma metastasis. We identified TRAIP as a key differentially expressed gene with prognostic significance in osteosarcoma. We demonstrated that TRAIP regulated the proliferation and invasion of osteosarcoma cells. In addition, we found that TRAIP promoted KANK1 polyubiquitination and subsequent degradation, downregulating IGFBP3 and activating the AKT pathway in osteosarcoma cells. These results support the critical role of the TRAIP/ KANK1/IGFBP3/AKT signaling axis in osteosarcoma progression and suggest that TRAIP may represent a promising therapeutic target for osteosarcoma. Cell Death and Disease (2021) 12:767 ; https://doi.org/10.1038/s41419-021-04057-0 INTRODUCTION mechanisms underlying osteosarcoma metastasis. We identified Mesenchymal stem cell-derived osteosarcoma is one of the most TRAIP as a differentially expressed gene (DEG) with prognostic and common primary malignancies in bones [1], and it is particularly diagnostic significance. We also found that TRAIP regulated the common in children and adolescents. With the recent progress in proliferation and invasion of osteosarcoma cells. -

RING-Type E3 Ligases: Master Manipulators of E2 Ubiquitin-Conjugating Enzymes and Ubiquitination☆
Biochimica et Biophysica Acta 1843 (2014) 47–60 Contents lists available at ScienceDirect Biochimica et Biophysica Acta journal homepage: www.elsevier.com/locate/bbamcr Review RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination☆ Meredith B. Metzger a,1, Jonathan N. Pruneda b,1, Rachel E. Klevit b,⁎, Allan M. Weissman a,⁎⁎ a Laboratory of Protein Dynamics and Signaling, Center for Cancer Research, National Cancer Institute, 1050 Boyles Street, Frederick, MD 21702, USA b Department of Biochemistry, Box 357350, University of Washington, Seattle, WA 98195, USA article info abstract Article history: RING finger domain and RING finger-like ubiquitin ligases (E3s), such as U-box proteins, constitute the vast Received 5 March 2013 majority of known E3s. RING-type E3s function together with ubiquitin-conjugating enzymes (E2s) to medi- Received in revised form 23 May 2013 ate ubiquitination and are implicated in numerous cellular processes. In part because of their importance in Accepted 29 May 2013 human physiology and disease, these proteins and their cellular functions represent an intense area of study. Available online 6 June 2013 Here we review recent advances in RING-type E3 recognition of substrates, their cellular regulation, and their varied architecture. Additionally, recent structural insights into RING-type E3 function, with a focus on im- Keywords: RING finger portant interactions with E2s and ubiquitin, are reviewed. This article is part of a Special Issue entitled: U-box Ubiquitin–Proteasome System. Guest Editors: Thomas Sommer and Dieter H. Wolf. Ubiquitin ligase (E3) Published by Elsevier B.V. Ubiquitin-conjugating enzyme (E2) Protein degradation Catalysis 1. -

Supplementary Figure S1 Functional Characterisation of Snmp:GFP
doi: 10.1038/nature06328 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE LEGENDS Figure S1 | Functional characterisation of SNMP fusion proteins. Dose-response curve for cVA in Or67d neurons of wild-type (Berlin), SNMP mutant (Or67d- GAL4/+;SNMP1/SNMP2), SNMP:GFP rescue (Or67d-GAL4/UAS- SNMP:GFP;SNMP1/SNMP2) and YFP(1):Or83b/SNMP:YFP(2) rescue (Or67d:GAL4,UAS-YFP(1):Or83b/UAS-SNMP:YFP(2);SNMP1,Or83b2/SNMP2,Or83b1 ) animals. Mean responses are plotted (± s.e.m; wild-type n=47, SNMP mutant n=46, SNMP:GFP rescue n=20; YFP(1):Or83b/SNMP:YFP(2) rescue n=22; ≤4 sensilla/animal, mixed genders). Wild-type and SNMP:GFP rescue responses to cVA are not significantly different (ANOVA; p>0.1175). YFP(1):Or83b/SNMP:YFP(2) rescue responses to cVA are highly significantly different from SNMP mutants and from wild- type (ANOVA; p<0.0001), indicating partial rescue. Figure S2 | Cell type-specific rescue of SNMP expression. a, Immunostaining for mCD8:GFP (anti-GFP, green) and LUSH (magenta) in LUSH-GAL4/UAS-mCD8:GFP animals reveals faithful recapitulation of endogenous expression by the LUSH-GAL4 driver. b, Two-colour RNA in situ hybridisation for SNMP (green) and Or67d (magenta) in antennal sections of wild-type, Or67d neuron SNMP rescue (Or67d-GAL4/UAS- SNMP;SNMP1/SNMP2) and support cell SNMP rescue (LUSH-GAL4/UAS- SNMP;SNMP1/SNMP2) animals. www.nature.com/nature 1 Benton et al., Figure S1 ) -1 wild-type 120 SNMP:GFP rescue 80 YFP(1):Or83b/SNMP:YFP(2) rescue 40 Corrected response (spikes s 0 SNMP-/- 0 0.1 1 10 100 cVA (%) www.nature.com/nature 2 Benton -

UBE2H Protein UBE2H Protein
Catalogue # Aliquot Size U223-30H-20 20 µg U223-30H-50 50 µg UBE2H Protein Full length recombinant protein expressed in E. coli cells Catalog # U223-30H Lot # J581 -2 Product Description Purity Recombinant full length human UBE2H was expressed in E. coli cells using an N-terminal His tag. The gene accession number is NM_003344 . The purity of UBE2H was determined Gene Aliases to be >95% by densitometry. Approx. MW 21 kDa . E2-20K; UBC8; UBCH; UBCH2 Formulation Recombinant protein stored in 50mM sodium phosphate, pH 7.0, 300mM NaCl, 150mM imidazole, 0.1mM PMSF, 0.25mM DTT, 25% glycerol. Storage and Stability Store product at –70 oC. For optimal storage, aliquot target into smaller quantities after centrifugation and store at recommended temperature. For most favorable performance, avoid repeated handling and multiple freeze/thaw cycles. Scientific Background UBE2H or ubiquitin-conjugating enzyme E2H is a member of the E2 ubiquitin-conjugating enzyme family which is located on chromosome 7 (1). UBE2H is known to act on histones and cytoskeletal proteins, both involved in the UBE2H Protein degenerative pathway of the motor neuron. UBE2H Full length recombinant protein expressed in E. coli cells expression is increased during erythroid differentiation of hCD34(+) cells. Tal1 transcription factor, which is essential Catalog Number U223-30H for the development of the hematopoietic system and Specific Lot Number J581-2 plays a role during definitive erythropoiesis in the adult, Purity >95% activates UBE2H expression, whereas Tal1 knock-down Concentration 0.1µg/ µl Stability 1yr At –70 oC from date of shipment reduced UBE2H expression and ubiquitin transfer activity Storage & Shipping Store product at –70 oC. -

Hippo and Sonic Hedgehog Signalling Pathway Modulation of Human Urothelial Tissue Homeostasis
Hippo and Sonic Hedgehog signalling pathway modulation of human urothelial tissue homeostasis Thomas Crighton PhD University of York Department of Biology November 2020 Abstract The urinary tract is lined by a barrier-forming, mitotically-quiescent urothelium, which retains the ability to regenerate following injury. Regulation of tissue homeostasis by Hippo and Sonic Hedgehog signalling has previously been implicated in various mammalian epithelia, but limited evidence exists as to their role in adult human urothelial physiology. Focussing on the Hippo pathway, the aims of this thesis were to characterise expression of said pathways in urothelium, determine what role the pathways have in regulating urothelial phenotype, and investigate whether the pathways are implicated in muscle-invasive bladder cancer (MIBC). These aims were assessed using a cell culture paradigm of Normal Human Urothelial (NHU) cells that can be manipulated in vitro to represent different differentiated phenotypes, alongside MIBC cell lines and The Cancer Genome Atlas resource. Transcriptomic analysis of NHU cells identified a significant induction of VGLL1, a poorly understood regulator of Hippo signalling, in differentiated cells. Activation of upstream transcription factors PPARγ and GATA3 and/or blockade of active EGFR/RAS/RAF/MEK/ERK signalling were identified as mechanisms which induce VGLL1 expression in NHU cells. Ectopic overexpression of VGLL1 in undifferentiated NHU cells and MIBC cell line T24 resulted in significantly reduced proliferation. Conversely, knockdown of VGLL1 in differentiated NHU cells significantly reduced barrier tightness in an unwounded state, while inhibiting regeneration and increasing cell cycle activation in scratch-wounded cultures. A signalling pathway previously observed to be inhibited by VGLL1 function, YAP/TAZ, was unaffected by VGLL1 manipulation. -

Supp Table 6.Pdf
Supplementary Table 6. Processes associated to the 2037 SCL candidate target genes ID Symbol Entrez Gene Name Process NM_178114 AMIGO2 adhesion molecule with Ig-like domain 2 adhesion NM_033474 ARVCF armadillo repeat gene deletes in velocardiofacial syndrome adhesion NM_027060 BTBD9 BTB (POZ) domain containing 9 adhesion NM_001039149 CD226 CD226 molecule adhesion NM_010581 CD47 CD47 molecule adhesion NM_023370 CDH23 cadherin-like 23 adhesion NM_207298 CERCAM cerebral endothelial cell adhesion molecule adhesion NM_021719 CLDN15 claudin 15 adhesion NM_009902 CLDN3 claudin 3 adhesion NM_008779 CNTN3 contactin 3 (plasmacytoma associated) adhesion NM_015734 COL5A1 collagen, type V, alpha 1 adhesion NM_007803 CTTN cortactin adhesion NM_009142 CX3CL1 chemokine (C-X3-C motif) ligand 1 adhesion NM_031174 DSCAM Down syndrome cell adhesion molecule adhesion NM_145158 EMILIN2 elastin microfibril interfacer 2 adhesion NM_001081286 FAT1 FAT tumor suppressor homolog 1 (Drosophila) adhesion NM_001080814 FAT3 FAT tumor suppressor homolog 3 (Drosophila) adhesion NM_153795 FERMT3 fermitin family homolog 3 (Drosophila) adhesion NM_010494 ICAM2 intercellular adhesion molecule 2 adhesion NM_023892 ICAM4 (includes EG:3386) intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)adhesion NM_001001979 MEGF10 multiple EGF-like-domains 10 adhesion NM_172522 MEGF11 multiple EGF-like-domains 11 adhesion NM_010739 MUC13 mucin 13, cell surface associated adhesion NM_013610 NINJ1 ninjurin 1 adhesion NM_016718 NINJ2 ninjurin 2 adhesion NM_172932 NLGN3 neuroligin -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -
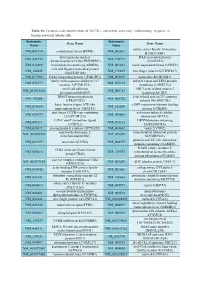
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973