Phylogenetic Relationships Among the Delphinid Cetaceans Based on Full Cytochrome B Sequences R
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Download Full Article in PDF Format
A new marine vertebrate assemblage from the Late Neogene Purisima Formation in Central California, part II: Pinnipeds and Cetaceans Robert W. BOESSENECKER Department of Geology, University of Otago, 360 Leith Walk, P.O. Box 56, Dunedin, 9054 (New Zealand) and Department of Earth Sciences, Montana State University 200 Traphagen Hall, Bozeman, MT, 59715 (USA) and University of California Museum of Paleontology 1101 Valley Life Sciences Building, Berkeley, CA, 94720 (USA) [email protected] Boessenecker R. W. 2013. — A new marine vertebrate assemblage from the Late Neogene Purisima Formation in Central California, part II: Pinnipeds and Cetaceans. Geodiversitas 35 (4): 815-940. http://dx.doi.org/g2013n4a5 ABSTRACT e newly discovered Upper Miocene to Upper Pliocene San Gregorio assem- blage of the Purisima Formation in Central California has yielded a diverse collection of 34 marine vertebrate taxa, including eight sharks, two bony fish, three marine birds (described in a previous study), and 21 marine mammals. Pinnipeds include the walrus Dusignathus sp., cf. D. seftoni, the fur seal Cal- lorhinus sp., cf. C. gilmorei, and indeterminate otariid bones. Baleen whales include dwarf mysticetes (Herpetocetus bramblei Whitmore & Barnes, 2008, Herpetocetus sp.), two right whales (cf. Eubalaena sp. 1, cf. Eubalaena sp. 2), at least three balaenopterids (“Balaenoptera” cortesi “var.” portisi Sacco, 1890, cf. Balaenoptera, Balaenopteridae gen. et sp. indet.) and a new species of rorqual (Balaenoptera bertae n. sp.) that exhibits a number of derived features that place it within the genus Balaenoptera. is new species of Balaenoptera is relatively small (estimated 61 cm bizygomatic width) and exhibits a comparatively nar- row vertex, an obliquely (but precipitously) sloping frontal adjacent to vertex, anteriorly directed and short zygomatic processes, and squamosal creases. -

Marine Mammal Conservation from Local to Global
MARINE MAMMAL CONSERVATION FROM LOCAL TO GLOBAL 29TH CONFERENCE OF THE EUROPEAN CETACEAN SOCIETY 23rd to 25th March, 2015 Intercontinental Hotel, St Julian’s Bay, MALTA USEFUL INFORMATION VENUE – INTERCONTIMENTAL MALTA HOTEL, ST JULIANS Conference Hall, Cettina De Cesare (CDC), is in hotel. Paranga Beach Club is on the water edge in St George’s Bay. 29th ECS Conference, Malta i USEFUL INFORMATION CONTACT NUMBERS Direct Dialling Code for Malta: +356 International Code (to make an overseas call): 00 Emergency number: 112 Police: 21 22 40 01 … 21 22 40 07 Mater-Dei Hospital (Malta): 25 45 00 00 Malta International Airport (General Inquiries): 21 24 96 00 Malta International Airport (Flight Information): 52 30 20 00 (each call: € 1.00) Passport Office: 21 22 22 86 WEBSITES Malta International Airport (note one ‘a’ between Malta and Airport!) Malta’s weather www.maltairport.com/weather Arrivals www.maltairport.com/arrivals Departures www.maltairport.com/departures Activities in Malta www.visitmalta.com 29th ECS Conference, Malta ii ACKNOWLEDGEMENTS HOSTED BY The Biological Conservation Research Foundation (BICREF) The NGO BICREF was set-up in 1998 to promote conservation research and awareness in Malta. For this purpose it welcomes Internships in Malta; the next call starts immediately after the ECS conference 2015 and to last till the end of summer 2015. Options for taking up courses or training in marine conservation biology, cetacean and fisheries research are also possible. Dr. Adriana Vella, Ph.D (Cantab.), founder of BICREF, is a conservation biologist with experience in mammal and marine conservation research at local and regional level. -

Ecological Factors Influencing Group Sizes of River Dolphins (Inia Geoffrensis and Sotalia Fluviatilis)
MARINE MAMMAL SCIENCE, **(*): ***–*** (*** 2011) C 2011 by the Society for Marine Mammalogy DOI: 10.1111/j.1748-7692.2011.00496.x Ecological factors influencing group sizes of river dolphins (Inia geoffrensis and Sotalia fluviatilis) CATALINA GOMEZ-SALAZAR Biology Department, Dalhousie University, Halifax, Nova Scotia B3H4J1, Canada and Foundation Omacha, Calle 86A No. 23–38, Bogota, Colombia E-mail: [email protected] FERNANDO TRUJILLO Foundation Omacha, Calle 86A No. 23–38, Bogota, Colombia HAL WHITEHEAD Biology Department, Dalhousie University, Halifax, Nova Scotia B3H4J1, Canada ABSTRACT Living in groups is usually driven by predation and competition for resources. River dolphins do not have natural predators but inhabit dynamic systems with predictable seasonal shifts. These ecological features may provide some insight into the forces driving group formation and help us to answer questions such as why river dolphins have some of the smallest group sizes of cetaceans, and why group sizes vary with time and place. We analyzed observations of group size for Inia and Sotalia over a 9 yr period. In the Amazon, largest group sizes occurred in main rivers and lakes, particularly during the low water season when resources are concentrated; smaller group sizes occurred in constricted waters (channels, tributaries, and confluences) that receive an influx of blackwaters that are poor in nutrients and sediments. In the Orinoco, the largest group sizes occurred during the transitional water season when the aquatic productivity increases. The largest group size of Inia occurred in the Orinoco location that contains the influx of two highly productive whitewater rivers. Flood pulses govern productivity and major biological factors of these river basins. -

EXTERNAL FEATURES of the DUSKY DOLPHIN Lagenorhynchus Obscurus (GRAY, 1828) from PERUVIAN WATERS Caracteristicas EXTERNAS DEL DE
Estud. Oceanol. 12: 37-53 1993 ISSN CL 0O71-173X EXTERNAL FEATURES OF THE DUSKY DOLPHIN Lagenorhynchus obscurus (GRAY, 1828) FROM PERUVIAN WATERS CARACTERisTICAS EXTERNAS DEL DELFiN OSCURO Lagenorhynchus obscurus (GRAY, 1828) DE AGUAS PERUANAS Koen Van Waerebeek Centro Peruano de Estudios Cetol6gicos (CEPEC), Asociaci6n de Ecologfa y Conservaci6n, Casilla 1536, Lima 18, Peru ABSTRACT Individual, sexual and developmental variation is quantified in the external morphology and colouration of the dusky dolphin Lagenorhynchus obscurus from Peruvian coastal waters. No significant difference in body length between sexes is found{p = 0.09) and, generally, little sexual dimorphism is present. However, males have a more anteriorly positioned genital slit and anus and their dorsal fin is more curved, has a broader base and a greater surface area than females Although the dorsal fin apparently serves as a secondary sexual character, the use of it for sexing free-ranging dusky dolphins is discouraged because of high overlap in values Relative growth in 25 body measurements is characterized for both sexes by multiplicative regression equations. The colouration pattern of the dorsal fin, flank patch, thoracic field, flipper stripe and possibly (x2, p = 008) the eye patch, are independent of maturity status. Flipper blaze and lower lip patch are less pigmented in juveniles than in adults No sexual dimorphism is found in the colour pattern The existence of a discrete "Fitzroy" colour form can not be confirmed from available data. Various cases of anomalous, piebald pigmentation are described, probably equivalent to so-called partial albinism Adult dusky dolphins from both SW Africa and New Zealand are 8-10 cm shorter than Peruvian specimens, supporting conclusions of separate populations from a recent skull variability study. -

THE CASE AGAINST Marine Mammals in Captivity Authors: Naomi A
s l a m m a y t T i M S N v I i A e G t A n i p E S r a A C a C E H n T M i THE CASE AGAINST Marine Mammals in Captivity The Humane Society of the United State s/ World Society for the Protection of Animals 2009 1 1 1 2 0 A M , n o t s o g B r o . 1 a 0 s 2 u - e a t i p s u S w , t e e r t S h t u o S 9 8 THE CASE AGAINST Marine Mammals in Captivity Authors: Naomi A. Rose, E.C.M. Parsons, and Richard Farinato, 4th edition Editors: Naomi A. Rose and Debra Firmani, 4th edition ©2009 The Humane Society of the United States and the World Society for the Protection of Animals. All rights reserved. ©2008 The HSUS. All rights reserved. Printed on recycled paper, acid free and elemental chlorine free, with soy-based ink. Cover: ©iStockphoto.com/Ying Ying Wong Overview n the debate over marine mammals in captivity, the of the natural environment. The truth is that marine mammals have evolved physically and behaviorally to survive these rigors. public display industry maintains that marine mammal For example, nearly every kind of marine mammal, from sea lion Iexhibits serve a valuable conservation function, people to dolphin, travels large distances daily in a search for food. In learn important information from seeing live animals, and captivity, natural feeding and foraging patterns are completely lost. -

Mammal Species Native to the USA and Canada for Which the MIL Has an Image (296) 31 July 2021
Mammal species native to the USA and Canada for which the MIL has an image (296) 31 July 2021 ARTIODACTYLA (includes CETACEA) (38) ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BALAENIDAE - bowheads and right whales 1. Balaena mysticetus – Bowhead Whale BALAENOPTERIDAE -rorqual whales 1. Balaenoptera acutorostrata – Common Minke Whale 2. Balaenoptera borealis - Sei Whale 3. Balaenoptera brydei - Bryde’s Whale 4. Balaenoptera musculus - Blue Whale 5. Balaenoptera physalus - Fin Whale 6. Eschrichtius robustus - Gray Whale 7. Megaptera novaeangliae - Humpback Whale BOVIDAE - cattle, sheep, goats, and antelopes 1. Bos bison - American Bison 2. Oreamnos americanus - Mountain Goat 3. Ovibos moschatus - Muskox 4. Ovis canadensis - Bighorn Sheep 5. Ovis dalli - Thinhorn Sheep CERVIDAE - deer 1. Alces alces - Moose 2. Cervus canadensis - Wapiti (Elk) 3. Odocoileus hemionus - Mule Deer 4. Odocoileus virginianus - White-tailed Deer 5. Rangifer tarandus -Caribou DELPHINIDAE - ocean dolphins 1. Delphinus delphis - Common Dolphin 2. Globicephala macrorhynchus - Short-finned Pilot Whale 3. Grampus griseus - Risso's Dolphin 4. Lagenorhynchus albirostris - White-beaked Dolphin 5. Lissodelphis borealis - Northern Right-whale Dolphin 6. Orcinus orca - Killer Whale 7. Peponocephala electra - Melon-headed Whale 8. Pseudorca crassidens - False Killer Whale 9. Sagmatias obliquidens - Pacific White-sided Dolphin 10. Stenella coeruleoalba - Striped Dolphin 11. Stenella frontalis – Atlantic Spotted Dolphin 12. Steno bredanensis - Rough-toothed Dolphin 13. Tursiops truncatus - Common Bottlenose Dolphin MONODONTIDAE - narwhals, belugas 1. Delphinapterus leucas - Beluga 2. Monodon monoceros - Narwhal PHOCOENIDAE - porpoises 1. Phocoena phocoena - Harbor Porpoise 2. Phocoenoides dalli - Dall’s Porpoise PHYSETERIDAE - sperm whales Physeter macrocephalus – Sperm Whale TAYASSUIDAE - peccaries Dicotyles tajacu - Collared Peccary CARNIVORA (48) CANIDAE - dogs 1. Canis latrans - Coyote 2. -

Extrapolating Cetacean Densities Beyond Surveyed Regions: Habitat
Journal of Biogeography (J. Biogeogr.) (2015) 42, 1267–1280 ORIGINAL Extrapolating cetacean densities beyond ARTICLE surveyed regions: habitat-based predictions in the circumtropical belt Laura Mannocci1*, Pascal Monestiez1,2,Jerome^ Spitz3,4 and Vincent Ridoux1,3 1Centre d’Etudes Biologiques de Chize et La ABSTRACT Rochelle, UMR 7372 Universite de La Aim Our knowledge of cetacean distributions is impeded by large data-gaps Rochelle-CNRS, La Rochelle F-17000, France, 2 worldwide, particularly at tropical latitudes. This study aims to (1) find generic INRA, UR0546, Unite Biostatistiques et Processus Spatiaux, Domaine Saint-Paul relationships between cetaceans and their habitats in a range of tropical waters, 84914, Avignon, France, 3Observatoire and (2) extrapolate cetacean densities in a circumtropical belt extending far PELAGIS, UMS 3462 Universite de La beyond surveyed regions. Rochelle-CNRS, Systemes d’Observation pour Location Pelagic, circumtropical. la Conservation des Mammiferes et des Oiseaux Marins, La Rochelle 17000, France, Methods Aerial surveys were conducted over three regions in the tropical 2 2 2 4Marine Mammal Research Unit, Fisheries Atlantic (132,000 km ), Indian (1.4 million km ) and Pacific (1.4 million km ) Center, University of British Columbia, oceans. Three cetacean guilds were studied (Delphininae, Globicephalinae and Vancouver, British Columbia V6T 1Z4, sperm and beaked whales). For each guild, a generalized additive model was Canada fitted using sightings recorded in all three regions and 14 candidate environ- mental predictors. Cetacean densities were tentatively extrapolated over a cir- cumtropical belt, excluding waters where environmental characteristics departed from those encountered in the surveyed regions. Results Each cetacean guild exhibited a relationship with the primary produc- tion and depth of the minimum dissolved oxygen concentration. -

Cetacea: Phocoenidae) from the Upper Part of the Horokaoshirarika Formation (Lower Pliocene), Numata Town, Hokkaido, Japan, and Its Phylogenetic Position
Palaeontologia Electronica palaeo-electronica.org A new skull of the fossil porpoise Numataphocoena yamashitai (Cetacea: Phocoenidae) from the upper part of the Horokaoshirarika Formation (lower Pliocene), Numata Town, Hokkaido, Japan, and its phylogenetic position Yoshihiro Tanaka and Hiroto Ichishima ABSTRACT An early Pliocene porpoise, Numataphocoena yamashitai from Hokkaido, Japan, is known from the holotype, a fairly well-preserved skeleton with an incomplete skull and a referred earbone. A new skull referred to Numataphocoena yamashitai found from almost the same locality as the holotype is interesting because it expands knowl- edge of skull morphology and improves the diagnosis of this taxon. Numataphocoena yamashitai differs from other phocoenids in having the characteristic feature in the maxilla associated with the posterior dorsal infraorbital foramen, narrower and sharper anterior part of the internal acoustic meatus, and a robust anterior process of the peri- otic. A new cladistic analysis places Numataphocoena yamashitai adjacent to Haboro- phocoena toyoshimai and Haborophocoena minutus, among a clade of early branching phocoenids, all of which are chronologically and geographically close to each other. The new skull is probably a younger individual because it is about 80% the size of that of the holotype and it shows closed but unfused sutures. Our description of this specimen helps to understand the intraspecies variation of the extinct species Numataphocoena yamashitai. Yoshihiro Tanaka. Numata Fossil Museum, 2-7-49, Minami 1, Numata Town, Hokkaido, 078-2225 Japan, [email protected] and Hokkaido University Museum, Kita 10, Nishi 8, Kita-ku, Sapporo, Hokkaido 060-0810 Japan Hiroto Ichishima. Fukui Prefectural Dinosaur Museum, Terao 51-11, Muroko, Katsuyama, Fukui 911-8601, Japan, [email protected] Key words: skull; Phocoenidae; phylogeny; maxillary terrace; ontogeny; intraspecies variation Submission: 22 March 2016 Acceptance: 20 October 2016 Tanaka, Yoshihiro and Ichishima, Hiroto. -

Taxonomic Status of the Genus Sotalia: Species Level Ranking for “Tucuxi” (Sotalia Fluviatilis) and “Costero” (Sotalia Guianensis) Dolphins
MARINE MAMMAL SCIENCE, **(*): ***–*** (*** 2007) C 2007 by the Society for Marine Mammalogy DOI: 10.1111/j.1748-7692.2007.00110.x TAXONOMIC STATUS OF THE GENUS SOTALIA: SPECIES LEVEL RANKING FOR “TUCUXI” (SOTALIA FLUVIATILIS) AND “COSTERO” (SOTALIA GUIANENSIS) DOLPHINS S. CABALLERO Laboratory of Molecular Ecology and Evolution, School of Biological Sciences, University of Auckland, Private Bag 92019, Auckland, New Zealand and Fundacion´ Omacha, Diagonal 86A #30–38, Bogota,´ Colombia F. TRUJILLO Fundacion´ Omacha, Diagonal 86A #30–38, Bogota,´ Colombia J. A. VIANNA Sala L3–244, Departamento de Biologia Geral, ICB, Universidad Federal de Minas Gerais, Avenida Antonio Carlos, 6627 C. P. 486, 31270–010 Belo Horizonte, Brazil and Escuela de Medicina Veterinaria, Facultad de Ecologia y Recursos Naturales, Universidad Andres Bello Republica 252, Santigo, Chile H. BARRIOS-GARRIDO Laboratorio de Sistematica´ de Invertebrados Acuaticos´ (LASIA), Postgrado en Ciencias Biologicas,´ Facultad Experimental de Ciencias,Universidad del Zulia, Avenida Universidad con prolongacion´ Avenida 5 de Julio, Sector Grano de Oro, Maracaibo, Venezuela M. G. MONTIEL Laboratorio de Ecologıa´ y Genetica´ de Poblaciones, Centro de Ecologıa,´ Instituto Venezolano de Investigaciones Cientıficas´ (IVIC), San Antonio de los Altos, Carretera Panamericana km 11, Altos de Pipe, Estado Miranda, Venezuela S. BELTRAN´ -PEDREROS Laboratorio de Zoologia,´ Colec¸ao˜ Zoologica´ Paulo Burheim, Centro Universitario´ Luterano de Manaus, Manaus, Brazil 1 2 MARINE MAMMAL SCIENCE, VOL. **, NO. **, 2007 M. MARMONTEL Sociedade Civil Mamiraua,´ Rua Augusto Correa No.1 Campus do Guama,´ Setor Professional, Guama,´ C. P. 8600, 66075–110 Belem,´ Brazil M. C. SANTOS Projeto Atlantis/Instituto de Biologia da Conservac¸ao,˜ Laboratorio´ de Biologia da Conservac¸ao˜ de Cetaceos,´ Departamento de Zoologia, Universidade Estadual Paulista (UNESP), Campus Rio Claro, Sao˜ Paulo, Brazil M. -

Morphometrics of the Dolphin Genus Lagenorhynchus: Deciphering A
Morphometrics of the dolphin genus Lagenorhynchus: deciphering a contested phylogeny Allison Galezo1,2 and Nicole Vollmer1,3 1 Department of Vertebrate Zoology, Smithsonian Institution National Museum of Natural History 2 Department of Biology, Georgetown University 3 NOAA National Systematics Laboratory Background Results & Analysis Discussion • Our morphological data support the hypothesis that Figure 1. Phenogram from cluster analysis of dolphin skull Figure 2. Species symbols key with sample sizes. measurements. Calculated using Euclidian distances and the genus Lagenorhynchus is not monophyletic, evident Height l La. acutus (24) p C. commersonii (5) a b c Ward’s method. from the separation in the phenogram of La. albirostris Height p C. eutropia (2) Recent phylogenetic studies1-7 have indicated that the Distance l La. albirostris (10) and La. acutus from the other Lagenorhynchus species, 0 2 4 6 8 p genus Lagenorhynchus, currently containing the species 0 2 4 6 8 l La. australis (7) C. heavisidii (1) u Li. borealis (11) and the mix of genera in the lowermost clade (Figure 1). L. obliquidensa, L. acutusb , L. albirostrisc, L. obscurusd , L. l La. obliquidens (28) p C. hectori (2) n Unknown (1) • Our results show that La. obscurus and La. obliquidens e f cruciger , and L. australis , is not monophyletic. These C. commersonii l La. obscurus (15) are very similar morphologically, which supports the C. commersonii species were originally grouped together because of C.C. cocommemmersoniirsonii C. commeC. hersoniictori hypothesis that they are closely related: they have C. commeC. hersoniictori similarities in external morphology and coloration, but C. commeC. hersoniictori Figure 3. -
Global Patterns in Marine Mammal Distributions
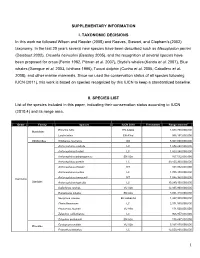
SUPPLEMENTARY INFORMATION I. TAXONOMIC DECISIONS In this work we followed Wilson and Reeder (2005) and Reeves, Stewart, and Clapham’s (2002) taxonomy. In the last 20 years several new species have been described such as Mesoplodon perrini (Dalebout 2002), Orcaella heinsohni (Beasley 2005), and the recognition of several species have been proposed for orcas (Perrin 1982, Pitman et al. 2007), Bryde's whales (Kanda et al. 2007), Blue whales (Garrigue et al. 2003, Ichihara 1996), Tucuxi dolphin (Cunha et al. 2005, Caballero et al. 2008), and other marine mammals. Since we used the conservation status of all species following IUCN (2011), this work is based on species recognized by this IUCN to keep a standardized baseline. II. SPECIES LIST List of the species included in this paper, indicating their conservation status according to IUCN (2010.4) and its range area. Order Family Species IUCN 2010 Freshwater Range area km2 Enhydra lutris EN A2abe 1,084,750,000,000 Mustelidae Lontra felina EN A3cd 996,197,000,000 Odobenidae Odobenus rosmarus DD 5,367,060,000,000 Arctocephalus australis LC 1,674,290,000,000 Arctocephalus forsteri LC 1,823,240,000,000 Arctocephalus galapagoensis EN A2a 167,512,000,000 Arctocephalus gazella LC 39,155,300,000,000 Arctocephalus philippii NT 163,932,000,000 Arctocephalus pusillus LC 1,705,430,000,000 Arctocephalus townsendi NT 1,045,950,000,000 Carnivora Otariidae Arctocephalus tropicalis LC 39,249,100,000,000 Callorhinus ursinus VU A2b 12,935,900,000,000 Eumetopias jubatus EN A2a 3,051,310,000,000 Neophoca cinerea -

Killer Whale) Orcinus Orca
AMERICAN CETACEAN SOCIETY FACT SHEET P.O. Box 1391 - San Pedro, CA 90733-1391 - (310) 548-6279 ORCA (Killer Whale) Orcinus orca CLASS: Mammalia ORDER: Cetacea SUBORDER: Odontoceti FAMILY: Delphinidae GENUS: Orcinus SPECIES: orca The orca, or killer whale, with its striking black and white coloring, is one of the best known of all the cetaceans. It has been extensively studied in the wild and is often the main attraction at many sea parks and aquaria. An odontocete, or toothed whale, the orca is known for being a carnivorous, fast and skillful hunter, with a complex social structure and a cosmopolitan distribution (orcas are found in all the oceans of the world). Sometimes called "the wolf of the sea", the orca can be a fierce hunter with well-organized hunting techniques, although there are no documented cases of killer whales attacking a human in the wild. PHYSICAL SHAPE The orca is a stout, streamlined animal. It has a round head that is tapered, with an indistinct beak and straight mouthline. COLOR The orca has a striking color pattern made up of well-defined areas of shiny black and cream or white. The dorsal (top) part of its body is black, with a pale white to gray "saddle" behind the dorsal fin. It has an oval, white eyepatch behind and above each eye. The chin, throat, central length of the ventral (underside) area, and undersides of the tail flukes are white. Each whale can be individually identified by its markings and by the shape of its saddle patch and dorsal fin.