11 Beacham FB107(2)
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Chapter 14. Northern Shelf Region
Chapter 14. Northern Shelf Region Queen Charlotte Sound, Hecate Strait, and Dixon canoes were almost as long as the ships of the early Spanish, Entrance form a continuous coastal seaway over the conti- and British explorers. The Haida also were gifted carvers nental shelfofthe Canadian west coast (Fig. 14.1). Except and produced a volume of art work which, like that of the for the broad lowlands along the northwest side ofHecate mainland tribes of the Kwaluutl and Tsimshian, is only Strait, the region is typified by a highly broken shoreline now becoming appreciated by the general public. of islands, isolated shoals, and countless embayments The first Europeans to sail the west coast of British which, during the last ice age, were covered by glaciers Columbia were Spaniards. Under the command of Juan that spread seaward from the mountainous terrain of the Perez they reached the vicinity of the Queen Charlotte mainland coast and the Queen Charlotte Islands. The Islands in 1774 before returning to a landfall at Nootka irregular countenance of the seaway is mirrored by its Sound on Vancouver Island. Quadra followed in 1775, bathymetry as re-entrant troughs cut landward between but it was not until after Cook’s voyage of 1778 with the shallow banks and broad shoals and extend into Hecate Resolution and Discovery that the white man, or “Yets- Strait from northern Graham Island. From an haida” (iron men) as the Haida called them, began to oceanographic point of view it is a hybrid region, similar explore in earnest the northern coastal waters. During his in many respects to the offshore waters but considerably sojourn at Nootka that year Cook had received a number modified by estuarine processes characteristic of the of soft, luxuriant sea otter furs which, after his death in protected inland coastal waters. -

British Columbia Regional Guide Cat
National Marine Weather Guide British Columbia Regional Guide Cat. No. En56-240/3-2015E-PDF 978-1-100-25953-6 Terms of Usage Information contained in this publication or product may be reproduced, in part or in whole, and by any means, for personal or public non-commercial purposes, without charge or further permission, unless otherwise specified. You are asked to: • Exercise due diligence in ensuring the accuracy of the materials reproduced; • Indicate both the complete title of the materials reproduced, as well as the author organization; and • Indicate that the reproduction is a copy of an official work that is published by the Government of Canada and that the reproduction has not been produced in affiliation with or with the endorsement of the Government of Canada. Commercial reproduction and distribution is prohibited except with written permission from the author. For more information, please contact Environment Canada’s Inquiry Centre at 1-800-668-6767 (in Canada only) or 819-997-2800 or email to [email protected]. Disclaimer: Her Majesty is not responsible for the accuracy or completeness of the information contained in the reproduced material. Her Majesty shall at all times be indemnified and held harmless against any and all claims whatsoever arising out of negligence or other fault in the use of the information contained in this publication or product. Photo credits Cover Left: Chris Gibbons Cover Center: Chris Gibbons Cover Right: Ed Goski Page I: Ed Goski Page II: top left - Chris Gibbons, top right - Matt MacDonald, bottom - André Besson Page VI: Chris Gibbons Page 1: Chris Gibbons Page 5: Lisa West Page 8: Matt MacDonald Page 13: André Besson Page 15: Chris Gibbons Page 42: Lisa West Page 49: Chris Gibbons Page 119: Lisa West Page 138: Matt MacDonald Page 142: Matt MacDonald Acknowledgments Without the works of Owen Lange, this chapter would not have been possible. -

RG 42 - Marine Branch
FINDING AID: 42-21 RECORD GROUP: RG 42 - Marine Branch SERIES: C-3 - Register of Wrecks and Casualties, Inland Waters DESCRIPTION: The finding aid is an incomplete list of Statement of Shipping Casualties Resulting in Total Loss. DATE: April 1998 LIST OF SHIPPING CASUALTIES RESULTING IN TOTAL LOSS IN BRITISH COLUMBIA COASTAL WATERS SINCE 1897 Port of Net Date Name of vessel Registry Register Nature of casualty O.N. Tonnage Place of casualty 18 9 7 Dec. - NAKUSP New Westminster, 831,83 Fire, B.C. Arrow Lake, B.C. 18 9 8 June ISKOOT Victoria, B.C. 356 Stranded, near Alaska July 1 MARQUIS OF DUFFERIN Vancouver, B.C. 629 Went to pieces while being towed, 4 miles off Carmanah Point, Vancouver Island, B.C. Sept.16 BARBARA BOSCOWITZ Victoria, B.C. 239 Stranded, Browning Island, Kitkatlah Inlet, B.C. Sept.27 PIONEER Victoria, B.C. 66 Missing, North Pacific Nov. 29 CITY OF AINSWORTH New Westminster, 193 Sprung a leak, B.C. Kootenay Lake, B.C. Nov. 29 STIRINE CHIEF Vancouver, B.C. Vessel parted her chains while being towed, Alaskan waters, North Pacific 18 9 9 Feb. 1 GREENWOOD Victoria, B.C. 89,77 Fire, laid up July 12 LOUISE Seaback, Wash. 167 Fire, Victoria Harbour, B.C. July 12 KATHLEEN Victoria, B.C. 590 Fire, Victoria Harbour, B.C. Sept.10 BON ACCORD New Westminster, 52 Fire, lying at wharf, B.C. New Westminster, B.C. Sept.10 GLADYS New Westminster, 211 Fire, lying at wharf, B.C. New Westminster, B.C. Sept.10 EDGAR New Westminster, 114 Fire, lying at wharf, B.C. -
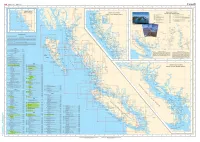
CHS Index Chart
Fisheries and Oceans Pi'lches et Oceans Canada Canada Canada ••• 137 ° 136 " 134" 133 ° 131° 129 ° ,.. 126 ° 125 " 124 " 123° 122° 119° 124 ° 118 ° GENERAL CHARTS CARTES GENERALES SMALL-CRAFT CHARTS REV ILLAG IGEDO LARGER SCALE CHARTS ISLAND CARTES POUR EMBARCATIONS CARTES A PLUS GRANDE ECHELLE 3050 Kootenay Lake and Rovet 75 000 3311 Sunshtno Coast- Vancouver Ha rbour lo/A 3052 Okanagan Lako so 000 Desolatoon Sound 40 000 3053 Shuswap Lake so 000 331:2 JerviS Intel ond/et Do•o latoon Sound 0 305S Waneta to /~ Hugh Keen leyside Dam 20 000 Vo,ous Scolo•JEche tle• vo"h• > z 3056 Hugh Koon loySido Dam to/A Burlon 40 000 3313 Gull Islands and Ad jacent Watotways/el les Vo1es Navigables Ad1acentes ~ ' 3057 Button IO/~ 1\rrowhood 40 000 Variou• Scale•/Echel le• vo"~"' 3058 Arrowhead lo/6 Rovo lotoko Go «m• 20 000 3488 Fro5er River/F I&uve Fraser, Cre•<ent l5land 3061 " "'"'on Lake and/ol Hamson R1ver '""'' to/~ Hon loon Mills 20 000 Harrison Lake 40 000 3469 Fraser Rovor/Fiouve Fr8set, Pattullo B"dgo Harrison R1ver 30 000 to/a Crescent Is land 20 000 Pitt River and/ot Poll l ak e 25 000 Stuaot L a~e (Not•howniP••rnd•qu o!) 50 000 54 " ~f--- ' ~ 0 "" I < ''"0' 't)Go iUn "?1- Cocoov• 3053 Foo ..o<o o l 0 "' GJ ,. Shu wap .•. Lake CANAOA !'; "'""""'' •·o~ d 130" 125° 120 " •5"omouo .,cocho Cceo> ,... ,. ' GJ ... ' <om l oops ~ DIXON E'N TRANCE' LEGENO/LEGENDE • •• • Scales smaller ttlan 1"40 000 Ectlellas plus petites qua 1:40 000 '' GJ Scales 1:40 000 and larger Ectlelles 1: 40 000 et plus grande& CHART SCALE Chart soale os the rat10 of one umt of d1stance 011 the cha" to the actual d.stance on the Earth's surface expressed on tho same unots. -

34 Haida Gwaii Is an Archipelago of More Than 200 Islands Located Be
4 THE ENVIRONMENT OF HAIDA GWAII 4.1 About the Islands Haida Gwaii is an archipelago of more than 200 islands located be- tween Vancouver Island and southeast Alaska, 50–30 km off the north- ern mainland coast of British Columbia (Figure .). The archipelago is scimitar shaped, and extends 250 km from north to south. Together, Graham and Moresby Islands constitute 90% of the nearly million hectares that comprise the archipelago. These two main islands are nar- rowly separated by Skidegate Inlet and Skidegate Channel. Other large islands in the archipelago include Louise, Lyell, and Burnaby Islands off the east coast of Moresby Island, and Kunghit Island to the south of Moresby Island. The landscape of the islands is extremely varied: it ranges from broad sand beaches and dunes on the east coast to the rocky, surf-battered west coast, and from muskeg lowlands on Graham Island to the rug- ged, strongly eroded Queen Charlotte Ranges of Moresby Island. The prevailing tranquil environment of Haida Gwaii is at times interrupted by dramatic natural processes—fierce storms, high-energy surf, floods, landslides, earthquakes, and windthrow—events that substantially modify the landscape and the ecosystems of the islands. In many ways, the archipelago is a microcosm of coastal British Columbia, but it has its own distinctive, and in some respects unique, natural and cultural heritage. The lands and waters of Haida Gwaii support a rich cultural heritage. Archaeological evidence indicates continuous human presence by the Haida and their ancestors dating back 0 000–3 000 years. Today, seven main communities are located on the east and north coasts of Graham Island and on the northeast tip of Moresby Island. -

Marine Distribution, Abundance, and Habitats of Marbled Murrelets in British Columbia
Chapter 29 Marine Distribution, Abundance, and Habitats of Marbled Murrelets in British Columbia Alan E. Burger1 Abstract: About 45,000-50,000 Marbled Murrelets (Brachyramphus birds) are extrapolations from relatively few data from 1972 marmoratus) breed in British Columbia, with some birds found in to 1982, mainly counts in high-density areas and transects most parts of the inshore coastline. A review of at-sea surveys at covering a small portion of coastline (Rodway and others 84 sites revealed major concentrations in summer in six areas. 1992). Between 1985 and 1993 many parts of the British Murrelets tend to leave these breeding areas in winter. Many Columbia coast were censused, usually by shoreline transects, murrelets overwinter in the Strait of Georgia and Puget Sound, but the wintering distribution is poorly known. Aggregations in sum- although methods and dates varied, making comparisons mer were associated with nearshore waters (<1 km from shore in and extrapolation difficult. Much of the 27,000 km of coastline exposed sites, but <3 km in sheltered waters), and tidal rapids and remains uncensused (fig. 1). narrows. Murrelets avoided deep fjord water. Several surveys showed Appendix 1 summarizes censuses for the core of the considerable daily and seasonal variation in densities, sometimes breeding season (1 May through 31 July). Murrelet densities linked with variable prey availability or local water temperatures. are given as birds per linear kilometer of transect. It was Anecdotal evidence suggests significant population declines in the necessary to convert density estimates from other units in Strait of Georgia, associated with heavy onshore logging in the several cases, and this was done in consultation with the early 1900s. -

Oceanography of the British Columbia Coast
CANADIAN SPECIAL PUBLICATION OF FISHERIES AND AQUATIC SCIENCES 56 DFO - L bra y / MPO B bliothèque Oceanography RI II I 111 II I I II 12038889 of the British Columbia Coast Cover photograph West Coast Moresby Island by Dr. Pat McLaren, Pacific Geoscience Centre, Sidney, B.C. CANADIAN SPECIAL PUBLICATION OF FISHERIES AND AQUATIC SCIENCES 56 Oceanography of the British Columbia Coast RICHARD E. THOMSON Department of Fisheries and Oceans Ocean Physics Division Institute of Ocean Sciences Sidney, British Columbia DEPARTMENT OF FISHERIES AND OCEANS Ottawa 1981 ©Minister of Supply and Services Canada 1981 Available from authorized bookstore agents and other bookstores, or you may send your prepaid order to the Canadian Government Publishing Centre Supply and Service Canada, Hull, Que. K1A 0S9 Make cheques or money orders payable in Canadian funds to the Receiver General for Canada A deposit copy of this publication is also available for reference in public librairies across Canada Canada: $19.95 Catalog No. FS41-31/56E ISBN 0-660-10978-6 Other countries:$23.95 ISSN 0706-6481 Prices subject to change without notice Printed in Canada Thorn Press Ltd. Correct citation for this publication: THOMSON, R. E. 1981. Oceanography of the British Columbia coast. Can. Spec. Publ. Fish. Aquat. Sci. 56: 291 p. for Justine and Karen Contents FOREWORD BACKGROUND INFORMATION Introduction Acknowledgments xi Abstract/Résumé xii PART I HISTORY AND NATURE OF THE COAST Chapter 5. Upwelling: Bringing Cold Water to the Surface Chapter 1. Historical Setting Causes of Upwelling 79 Origin of the Oceans 1 Localized Effects 82 Drifting Continents 2 Climate 83 Evolution of the Coast 6 Fishing Grounds 83 Early Exploration 9 El Nifio 83 Chapter 2. -

Of 38 IPHC Contracting Party Report: Canada DATE: 18/DEC/2020
IPHC-2020-AM096-NR01 Rev_1 IPHC Contracting Party Report: Canada DATE: 18/DEC/2020, REVISED 31/JAN/2020 CONTRACTING PARTY: CANADA AGENCY: Fisheries and Oceans Canada, Adam Keizer, Regional Manager, Groundfish, [email protected] Province of British Columbia, Minister of Agriculture Mike Turner, Senior Manager, Intergovernmental Relations, Fisheries, and Aquaculture, [email protected] Kevin Romanin, Senior Seafood Analyst, [email protected] FISHERY SECTOR/S All IPHC REGULATORY AREA/S IPHC Regulatory Area 2B (Canada: British Columbia) DISCUSSION Each year Fisheries and Oceans Canada provides harvest opportunities to First Nations for food, social and ceremonial (FSC) purposes (or domestic purposes for First Nations with modern treaties), and the commercial and recreational fisheries. First Nations, recreational, and commercial fisheries on the Pacific coast of Canada have long harvested groundfish. Groundfish serve as a source of food, they provide jobs, income, and enjoyment for individuals, businesses, and coastal communities and they play key roles in natural ecosystems. The B.C. Ministry of Agriculture is responsible for collection and reporting of data and statistics for the agri-food sector. An important part of that mandate is to analyze the impact of various sectors, including fisheries and seafood to the broader provincial economy. B.C. commercially harvests and reports on over 25 wild fisheries including Pacific halibut which is within B.C.’s top most valuable wild fishery commodities. Indigenous fisheries In the 1990 Sparrow decision, the Supreme Court of Canada found that where an Indigenous group has an Indigenous right to fish for food, social, and ceremonial (FSC) purposes, it takes priority, after conservation, over other uses of the resource. -

2009 Post Season Review Salmon North Coast Areas 1
2009 POST SEASON REVIEW SALMON NORTH COAST AREAS 1-6 & CENTRAL COAST AREAS 7-10 FISHERIES AND OCEANS CANADA 2009 POST SEASON REVIEW Table of Contents Table of Contents ..........................................................................................................................2-3 2009 Expectations and Results for Areas 1-6..................................................................................4-7 Statistical Area Map ......................................................................................................................8 Area 1 Sub-district Map............................................................................................................................9 2009 Post Season Summary and Assessment..................................................................................10-12 2009 Stream Escapements..............................................................................................................13 2009 FSC Catch Summary.............................................................................................................14 Area 2 East/West Sub-district Map............................................................................................................................15 2009 Post Season Summary and Assessment 2 East........................................................................16-18 2009 Commercial Net Catch ..........................................................................................................19 2009 Stream Escapements 2 East -

2016 IPHC Annual Meeting
2015 Canadian Recreational Fishery Halibut Catch Report January 11, 2016 Report Prepared for the International Pacific Halibut Commission (IPHC) Submitted by: Fisheries and Oceans Canada (DFO) 401 Burrard St. Vancouver, British Columbia Canada 0 Table of Contents 1. SUMMARY .............................................................................................................................. 3 1.1. Harvest ............................................................................................................................ 3 Table 1. Estimated Harvest in Pieces and Pounds by Regional Area ............................................................................ 3 Figure 1. Percentage of Halibut pieces harvested in pieces and weight by Regional Area. ........................................ 3 1.2. Biological Samples ........................................................................................................... 3 Table 2. Number of Halibut Biologically Sampled by Area and Source ........................................................................ 4 Figure 3. Halibut sampled for length or weight by area and the samples by source ................................................... 4 1.3. Fishery Logistics ............................................................................................................... 4 2. MANAGEMENT, MONITORING AND POLICY DEVELOPMENT ................................................... 5 2.1. Recreational Fishery Management Plan ......................................................................... -

Chapter 4 Seasonal Weather and Local Effects
BC-E 11/12/05 11:28 PM Page 75 LAKP-British Columbia 75 Chapter 4 Seasonal Weather and Local Effects Introduction 10,000 FT 7000 FT 5000 FT 3000 FT 2000 FT 1500 FT 1000 FT WATSON LAKE 600 FT 300 FT DEASE LAKE 0 SEA LEVEL FORT NELSON WARE INGENIKA MASSET PRINCE RUPERT TERRACE SANDSPIT SMITHERS FORT ST JOHN MACKENZIE BELLA BELLA PRINCE GEORGE PORT HARDY PUNTZI MOUNTAIN WILLAMS LAKE VALEMOUNT CAMPBELL RIVER COMOX TOFINO KAMLOOPS GOLDEN LYTTON NANAIMO VERNON KELOWNA FAIRMONT VICTORIA PENTICTON CASTLEGAR CRANBROOK Map 4-1 - Topography of GFACN31 Domain This chapter is devoted to local weather hazards and effects observed in the GFACN31 area of responsibility. After extensive discussions with weather forecasters, FSS personnel, pilots and dispatchers, the most common and verifiable hazards are listed. BC-E 11/12/05 11:28 PM Page 76 76 CHAPTER FOUR Most weather hazards are described in symbols on the many maps along with a brief textual description located beneath it. In other cases, the weather phenomena are better described in words. Table 3 (page 74 and 207) provides a legend for the various symbols used throughout the local weather sections. South Coast 10,000 FT 7000 FT 5000 FT 3000 FT PORT HARDY 2000 FT 1500 FT 1000 FT 600 FT 300 FT 0 SEA LEVEL CAMPBELL RIVER COMOX PEMBERTON TOFINO VANCOUVER HOPE NANAIMO ABBOTSFORD VICTORIA Map 4-2 - South Coast For most of the year, the winds over the South Coast of BC are predominately from the southwest to west. During the summer, however, the Pacific High builds north- ward over the offshore waters altering the winds to more of a north to northwest flow. -

Lode-Gold Deposits
BRITISH COLUMBIA DEPARTMENT OP MINES Hon. 15’. A. MCKE~UZIE,Minister. ROBT.DUNN, Deputy Xinister. J. D. CALLOVAY,Provincial Mineralogist. J. Droxso~,Chief Inspector of ?!Lines. D. E. WHITTAICER, ProrincialAssayer. BULLETIN No. 1, 1932 LODE-GOLD DEPOSITS OF BRITISlX COLUWBIIA COMPILED BY JOHN D. GALLOWAY, Provincial Mineralogist. PRINTED BY AUTEORlTY OB ‘lHF LEQISLATITE ASSEMBLY. To the Hoa. W.A. NcIfenzie, Ministar of Mines, Victoria, B.C. SIR-I beg to submit herewitha special bulletin on Lode-gold Depositsof British Columhia. This hulletin has been issued to supply the keen demand for infonuation regarding lode gold in the Province. The potentialities for successful sold-mining in the Province are greater than has been generally recognized and it is believed that the comgilation of all material information in one publication mill stimulate the exploitation of the latent gold resources of the Province. The reports by the Resident Engineerson their respecti7.e districts outlint; attraetiw opluxtnni- ties for capital in the opcning-up of known progerties, and give much information regarding likely arcas for the gold prospector. I hare the honour to he, Sir, Your obedient sen-ant, JOIIN D. GALLOWAY, PvovinciaZ Mineralogist. Bureau of .Wines, Victoria, KC.,Februwy Ist, 193%. LODE-GOLD DEPOSITS OF BRITISH COLUMBIA. - GENERAL SUMMARY. BY JOHND. GALLOTVAY,PitOSIHCIhL &~IKRXALOGIST. INTRODUCTION. Owing to the conditions attending silver and base-metal mining at the present time, renewed interest is being taken all overthe world in gold-mining, and this is ntostnoticeable in this Province.Lode-mining inBritish Columbia begxn with lode-gold productionfrom Various districts. Ic'or many years past, however, gold-mining has not becn as im))ortant as lead, copper, zinc, and silver mining in the Prouince.