Activation of Proneuronal Transcription Factor Ascl1 in Maternal Liver Ensures A
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Prospective Isolation of NKX2-1–Expressing Human Lung Progenitors Derived from Pluripotent Stem Cells
The Journal of Clinical Investigation RESEARCH ARTICLE Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells Finn Hawkins,1,2 Philipp Kramer,3 Anjali Jacob,1,2 Ian Driver,4 Dylan C. Thomas,1 Katherine B. McCauley,1,2 Nicholas Skvir,1 Ana M. Crane,3 Anita A. Kurmann,1,5 Anthony N. Hollenberg,5 Sinead Nguyen,1 Brandon G. Wong,6 Ahmad S. Khalil,6,7 Sarah X.L. Huang,3,8 Susan Guttentag,9 Jason R. Rock,4 John M. Shannon,10 Brian R. Davis,3 and Darrell N. Kotton1,2 2 1Center for Regenerative Medicine, and The Pulmonary Center and Department of Medicine, Boston University School of Medicine, Boston, Massachusetts, USA. 3Center for Stem Cell and Regenerative Medicine, Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center, Houston, Texas, USA. 4Department of Anatomy, UCSF, San Francisco, California, USA. 5Division of Endocrinology, Diabetes and Metabolism, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, Massachusetts, USA. 6Department of Biomedical Engineering and Biological Design Center, Boston University, Boston, Massachusetts, USA. 7Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA. 8Columbia Center for Translational Immunology & Columbia Center for Human Development, Columbia University Medical Center, New York, New York, USA. 9Department of Pediatrics, Monroe Carell Jr. Children’s Hospital, Vanderbilt University, Nashville, Tennessee, USA. 10Division of Pulmonary Biology, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA. It has been postulated that during human fetal development, all cells of the lung epithelium derive from embryonic, endodermal, NK2 homeobox 1–expressing (NKX2-1+) precursor cells. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

(12) Patent Application Publication (10) Pub. No.: US 2009/0111743 A1 Takahashi (43) Pub
US 200901 11743A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0111743 A1 Takahashi (43) Pub. Date: Apr. 30, 2009 (54) CYSTEINE-BRANCHED HEPARIN-BINDING Related U.S. Application Data GROWTH FACTOR ANALOGS (60) Provisional application No. 60/656,713, filed on Feb. 25, 2005. (75) Inventor: Kazuyuki Takahashi, O O Germantown, MD (US) Publication Classification (51) Int. Cl. Correspondence Address: A638/08C07K 700 30.82006.O1 201 THIRD STREET, N.W., SUITE 1340 A638/16 (2006.01) ALBUQUEROUE, NM 87102 (US) A638/10 (2006.01) A638/00 (2006.01) (73) Assignee: BioSurface Engineering (52) U.S. Cl. ........... 514/12:530/330; 530/329; 530/328; Technologies, Inc., Rockville, MD 530/327: 530/326; 530/325; 530/324; 514/17; (US) 514/16; 514/15: 514/14: 514/13 (57) ABSTRACT (21) Appl. No.: 11/362,137 The present invention provides a heparin binding growth factor analog of any of formula I-VIII and methods and uses (22) Filed: Feb. 23, 2006 thereof. US 2009/011 1743 A1 Apr. 30, 2009 CYSTENE-BRANCHED HEPARN-BINDING 0007 HBGFs useful in prevention or therapy of a wide GROWTH FACTOR ANALOGS range of diseases and disorders may be purified from natural sources or produced by recombinant DNA methods, however, CROSS-REFERENCE TO RELATED Such preparations are expensive and generally difficult to APPLICATIONS prepare. 0008. Some efforts have been made to generate heparin 0001. This application claims priority to and the benefit of binding growth factor analogs. For example, natural PDGF the filing of U.S. Provisional Patent Application Ser. -

Angiocrine Endothelium: from Physiology to Cancer Jennifer Pasquier1,2*, Pegah Ghiabi2, Lotf Chouchane3,4,5, Kais Razzouk1, Shahin Rafi3 and Arash Rafi1,2,3
Pasquier et al. J Transl Med (2020) 18:52 https://doi.org/10.1186/s12967-020-02244-9 Journal of Translational Medicine REVIEW Open Access Angiocrine endothelium: from physiology to cancer Jennifer Pasquier1,2*, Pegah Ghiabi2, Lotf Chouchane3,4,5, Kais Razzouk1, Shahin Rafi3 and Arash Rafi1,2,3 Abstract The concept of cancer as a cell-autonomous disease has been challenged by the wealth of knowledge gathered in the past decades on the importance of tumor microenvironment (TM) in cancer progression and metastasis. The sig- nifcance of endothelial cells (ECs) in this scenario was initially attributed to their role in vasculogenesis and angiogen- esis that is critical for tumor initiation and growth. Nevertheless, the identifcation of endothelial-derived angiocrine factors illustrated an alternative non-angiogenic function of ECs contributing to both physiological and pathological tissue development. Gene expression profling studies have demonstrated distinctive expression patterns in tumor- associated endothelial cells that imply a bilateral crosstalk between tumor and its endothelium. Recently, some of the molecular determinants of this reciprocal interaction have been identifed which are considered as potential targets for developing novel anti-angiocrine therapeutic strategies. Keywords: Angiocrine, Endothelium, Cancer, Cancer microenvironment, Angiogenesis Introduction of blood vessels in initiation of tumor growth and stated Metastatic disease accounts for about 90% of patient that in the absence of such angiogenesis, tumors can- mortality. Te difculty in controlling and eradicating not expand their mass or display a metastatic phenotype metastasis might be related to the heterotypic interaction [7]. Based on this theory, many investigators assumed of tumor and its microenvironment [1]. -

Type of the Paper (Article
Table S1. Gene expression of pro-angiogenic factors in tumor lymph nodes of Ibtk+/+Eµ-myc and Ibtk+/-Eµ-myc mice. Fold p- Symbol Gene change value 0,007 Akt1 Thymoma viral proto-oncogene 1 1,8967 061 0,929 Ang Angiogenin, ribonuclease, RNase A family, 5 1,1159 481 0,000 Angpt1 Angiopoietin 1 4,3916 117 0,461 Angpt2 Angiopoietin 2 0,7478 625 0,258 Anpep Alanyl (membrane) aminopeptidase 1,1015 737 0,000 Bai1 Brain-specific angiogenesis inhibitor 1 4,0927 202 0,001 Ccl11 Chemokine (C-C motif) ligand 11 3,1381 149 0,000 Ccl2 Chemokine (C-C motif) ligand 2 2,8407 298 0,000 Cdh5 Cadherin 5 2,5849 744 0,000 Col18a1 Collagen, type XVIII, alpha 1 3,8568 388 0,003 Col4a3 Collagen, type IV, alpha 3 2,9031 327 0,000 Csf3 Colony stimulating factor 3 (granulocyte) 4,3332 258 0,693 Ctgf Connective tissue growth factor 1,0195 88 0,000 Cxcl1 Chemokine (C-X-C motif) ligand 1 2,67 21 0,067 Cxcl2 Chemokine (C-X-C motif) ligand 2 0,7507 631 0,000 Cxcl5 Chemokine (C-X-C motif) ligand 5 3,921 328 0,000 Edn1 Endothelin 1 3,9931 042 0,001 Efna1 Ephrin A1 1,6449 601 0,002 Efnb2 Ephrin B2 2,8858 042 0,000 Egf Epidermal growth factor 1,726 51 0,000 Eng Endoglin 0,2309 467 0,000 Epas1 Endothelial PAS domain protein 1 2,8421 764 0,000 Ephb4 Eph receptor B4 3,6334 035 V-erb-b2 erythroblastic leukemia viral oncogene homolog 2, 0,000 Erbb2 3,9377 neuro/glioblastoma derived oncogene homolog (avian) 024 0,000 F2 Coagulation factor II 3,8295 239 1 0,000 F3 Coagulation factor III 4,4195 293 0,002 Fgf1 Fibroblast growth factor 1 2,8198 748 0,000 Fgf2 Fibroblast growth factor -
Targeting Fibroblast Growth Factor Pathways in Endometrial Cancer
Curr Probl Cancer 41 (2017) 37–47 Contents lists available at ScienceDirect Curr Probl Cancer journal homepage: www.elsevier.com/locate/cpcancer Targeting fibroblast growth factor pathways in endometrial cancer Boris Winterhoff, MDa, Gottfried E. Konecny, MDb,* article info abstract Novel treatments that improve outcomes for patients with Keywords: recurrent or metastatic endometrial cancer (EC) remain an Endometrial cancer unmet need. Aberrant signaling by fibroblast growth factors Fibroblast growth factor (FGFs) and FGF receptors (FGFRs) has been implicated in Angiogenesis several human cancers. Activating mutations in FGFR2 have been found in up to 16% of ECs, suggesting an opportunity for targeted therapy. This review summarizes the role of the FGF pathway in angiogenesis and EC, and provides an overview of FGFR-targeted therapies under clinical develop- ment for the treatment of EC. & 2017 The Authors. Published by Elsevier Inc. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/). Introduction Endometrial cancer (EC) is the most common gynecologic cancer, with 54,870 new cases and 10,170 related deaths estimated for the United States in 2015.1 Localized disease is often curable with surgery and the 5-year relative survival rate for patients diagnosed with early-stage EC is high.1-3 However, for patients who present with metastatic disease or experience a recurrence, prognosis is poor.1-3 Response of metastatic EC to standard therapies (eg, adjuvant radiotherapy, brachytherapy, or chemotherapy) is limited and overall survival for most patients with recurrent or metastatic disease is r1year.2-4 Thus, novel treatment options for this disease remain an unmet need. -

Investigating Hypoxic Tumor Physiology Through Gene Expression Patterns
Oncogene (2003) 22, 5907–5914 & 2003 Nature Publishing Group All rights reserved 0950-9232/03 $25.00 www.nature.com/onc Investigating hypoxic tumor physiology through gene expression patterns Nicholas C Denko*,1, Lucrezia A Fontana1, Karen M Hudson1, Patrick D Sutphin1, Soumya Raychaudhuri2, Russ Altman2 and Amato J Giaccia1 1Division of Radiation and Cancer Biology, Department of Radiation Oncology, Stanford University School of Medicine, Stanford, CA 94305, USA; 2Department of Genetics, Stanford University School of Medicine, Stanford, CA 94305, USA Clinical evidence shows that tumor hypoxia is an relyupon oxygen-dependent energyproduction suffer independent prognostic indicator of poor patient outcome. when oxygen falls below a certain threshold level. Hypoxic tumors have altered physiologic processes, However, too much oxygen can also result in the including increased regions of angiogenesis, increased generation of toxic, damaging radical byproducts. The local invasion, increased distant metastasis and altered net effect of these opposing pressures necessitates apoptotic programs. Since hypoxia is a potent controller the evolution of molecular mechanisms to regulate the of gene expression, identifying hypoxia-regulated genes is cellular availabilityand usage of oxygen. a means to investigate the molecular response to hypoxic The response to low-oxygen conditions has been stress. Traditional experimental approaches have identi- identified in both procaryotes and eucaryotes. Although fied physiologic changes in hypoxic cells. Recent studies the mechanisms for gene regulation in response to low- have identified hypoxia-responsive genes that may define oxygen conditions are diverse, parallel systems have the mechanism(s) underlying these physiologic changes. evolved in different organisms that are transcriptionally For example, the regulation of glycolytic genes by responsive to low-oxygen. -
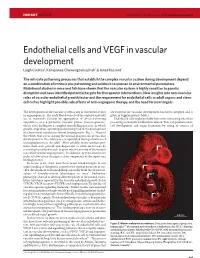
Endothelial Cells and VEGF in Vascular Development Leigh Coultas1, Kallayanee Chawengsaksophak1 & Janet Rossant1
Insight Rossant p7-15 6/12/05 10:23 AM Page 9377 INSIGHT REVIEW NATURE|Vol 438|15 December 2005|doi:10.1038/nature04479 Endothelial cells and VEGF in vascular development Leigh Coultas1, Kallayanee Chawengsaksophak1 & Janet Rossant1 The intricate patterning processes that establish the complex vascular system during development depend on a combination of intrinsic pre-patterning and extrinsic responses to environmental parameters. Mutational studies in mice and fish have shown that the vascular system is highly sensitive to genetic disruption and have identified potential targets for therapeutic interventions. New insights into non-vascular roles of vascular endothelial growth factor and the requirement for endothelial cells in adult organs and stem- cell niches highlight possible side effects of anti-angiogenic therapy and the need for new targets. The development of the vascular system is one of the earliest events are essential for vascular development has been compiled and is in organogenesis. The early blood vessels of the embryo and yolk given in Supplementary Table 1. sac in mammals develop by aggregation of de-novo-forming Endothelial cells might probably have more interesting roles than angioblasts into a primitive vascular plexus (vasculogenesis), just acting as channels for blood circulation. They can promote stem- which then undergoes a complex remodelling process, in which cell development and organ formation by acting as sources of growth, migration, sprouting and pruning lead to the development of a functional circulatory system (angiogenesis; Fig. 1). Many of the events that occur during the normal progression of vascular development in the embryo are recapitulated during situations of neoangiogenesis in the adult1. -

Vascular Endothelial Growth Factor (VEGF) Signaling in Tumor Progression Robert Roskoski Jr
Critical Reviews in Oncology/Hematology 62 (2007) 179–213 Vascular endothelial growth factor (VEGF) signaling in tumor progression Robert Roskoski Jr. ∗ Blue Ridge Institute for Medical Research, 3754 Brevard Road, Suite 116A, Box 19, Horse Shoe, NC 28742, USA Accepted 29 January 2007 Contents 1. Vasculogenesis and angiogenesis....................................................................................... 180 1.1. Definitions .................................................................................................... 180 1.2. Physiological and non-physiological angiogenesis ................................................................. 181 1.3. Activators and inhibitors of angiogenesis ......................................................................... 181 1.4. Sprouting and non-sprouting angiogenesis ........................................................................ 181 1.5. Tumor vessel morphology....................................................................................... 183 2. The vascular endothelial growth factor (VEGF) family ................................................................... 183 3. Properties and expression of the VEGF family .......................................................................... 183 3.1. VEGF-A ...................................................................................................... 183 3.2. VEGF-B ...................................................................................................... 185 3.3. VEGF-C ..................................................................................................... -

E Proteins Sharpen Neurogenesis by Modulating Proneural Bhlh
RESEARCH ARTICLE E proteins sharpen neurogenesis by modulating proneural bHLH transcription factors’ activity in an E-box-dependent manner Gwenvael Le Dre´ au1†*, Rene´ Escalona1†‡, Raquel Fueyo2, Antonio Herrera1, Juan D Martı´nez1, Susana Usieto1, Anghara Menendez3, Sebastian Pons3, Marian A Martinez-Balbas2, Elisa Marti1 1Department of Developmental Biology, Instituto de Biologı´a Molecular de Barcelona, Barcelona, Spain; 2Department of Molecular Genomics, Instituto de Biologı´a Molecular de Barcelona, Barcelona, Spain; 3Department of Cell Biology, Instituto de Biologı´a Molecular de Barcelona, Barcelona, Spain Abstract Class II HLH proteins heterodimerize with class I HLH/E proteins to regulate transcription. Here, we show that E proteins sharpen neurogenesis by adjusting the neurogenic strength of the distinct proneural proteins. We find that inhibiting BMP signaling or its target ID2 in *For correspondence: the chick embryo spinal cord, impairs the neuronal production from progenitors expressing [email protected] ATOH1/ASCL1, but less severely that from progenitors expressing NEUROG1/2/PTF1a. We show this context-dependent response to result from the differential modulation of proneural proteins’ †These authors contributed activity by E proteins. E proteins synergize with proneural proteins when acting on CAGSTG motifs, equally to this work thereby facilitating the activity of ASCL1/ATOH1 which preferentially bind to such motifs. Present address: Conversely, E proteins restrict the neurogenic strength of NEUROG1/2 by directly inhibiting their ‡ Departamento de Embriologı´a, preferential binding to CADATG motifs. Since we find this mechanism to be conserved in Facultad de Medicina, corticogenesis, we propose this differential co-operation of E proteins with proneural proteins as a Universidad Nacional Auto´noma novel though general feature of their mechanism of action. -
Figure S1. Reverse Transcription‑Quantitative PCR Analysis of ETV5 Mrna Expression Levels in Parental and ETV5 Stable Transfectants
Figure S1. Reverse transcription‑quantitative PCR analysis of ETV5 mRNA expression levels in parental and ETV5 stable transfectants. (A) Hec1a and Hec1a‑ETV5 EC cell lines; (B) Ishikawa and Ishikawa‑ETV5 EC cell lines. **P<0.005, unpaired Student's t‑test. EC, endometrial cancer; ETV5, ETS variant transcription factor 5. Figure S2. Survival analysis of sample clusters 1‑4. Kaplan Meier graphs for (A) recurrence‑free and (B) overall survival. Survival curves were constructed using the Kaplan‑Meier method, and differences between sample cluster curves were analyzed by log‑rank test. Figure S3. ROC analysis of hub genes. For each gene, ROC curve (left) and mRNA expression levels (right) in control (n=35) and tumor (n=545) samples from The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer cohort are shown. mRNA levels are expressed as Log2(x+1), where ‘x’ is the RSEM normalized expression value. ROC, receiver operating characteristic. Table SI. Clinicopathological characteristics of the GSE17025 dataset. Characteristic n % Atrophic endometrium 12 (postmenopausal) (Control group) Tumor stage I 91 100 Histology Endometrioid adenocarcinoma 79 86.81 Papillary serous 12 13.19 Histological grade Grade 1 30 32.97 Grade 2 36 39.56 Grade 3 25 27.47 Myometrial invasiona Superficial (<50%) 67 74.44 Deep (>50%) 23 25.56 aMyometrial invasion information was available for 90 of 91 tumor samples. Table SII. Clinicopathological characteristics of The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer dataset. Characteristic n % Solid tissue normal 16 Tumor samples Stagea I 226 68.278 II 19 5.740 III 70 21.148 IV 16 4.834 Histology Endometrioid 271 81.381 Mixed 10 3.003 Serous 52 15.616 Histological grade Grade 1 78 23.423 Grade 2 91 27.327 Grade 3 164 49.249 Molecular subtypeb POLE 17 7.328 MSI 65 28.017 CN Low 90 38.793 CN High 60 25.862 CN, copy number; MSI, microsatellite instability; POLE, DNA polymerase ε. -

Transcription Factor ATF5 Is Required for Terminal Differentiation and Survival of Olfactory Sensory Neurons
Transcription factor ATF5 is required for terminal differentiation and survival of olfactory sensory neurons Shu-Zong Wanga,b, Jianhong Oub, Lihua J. Zhub,c, and Michael R. Greena,b,1 aHoward Hughes Medical Institute, bPrograms in Gene Function and Expression and Molecular Medicine, and cProgram in Bioinformatics and Integrative Biology, University of Massachusetts Medical School, Worcester, MA 01605 Edited by Solomon H. Snyder, The Johns Hopkins University School of Medicine, Baltimore, MD, and approved September 27, 2012 (received for review June 21, 2012) Activating transcription factor 5 (ATF5) is a member of the ATF/cAMP not been determined. Here we address this issue by deriving and −/− response element-binding family of transcription factors, which characterizing Atf5 mice. compose a large group of basic region leucine zipper proteins whose members mediate diverse transcriptional regulatory functions. Results −/− ATF5 has a well-established prosurvival activity and has been found Derivation of Atf5 Mice. To investigate the physiological role of −/− to be overexpressed in several human cancers, in particular glioblas- ATF5, we derived Atf5 mice. The gene-targeting strategy in- toma. However, the role(s) of ATF5 in development and normal volved replacing almost the entire coding region of Atf5 with a physiology are unknown. Here we address this issue by deriving LacZ-Neo cassette, such that the LacZ gene is under the control and characterizing homozygous Atf5 knockout mice. We find that of the endogenous Atf5 promoter (Fig. S1A). The genomic struc- +/+ +/− −/− Atf5−/− pups die neonatally, which, as explained below, is consis- ture of the Atf5 locus in Atf5 , Atf5 ,andAtf5 mice was fi tent with an olfactory defect resulting in a competitive suckling con rmed by Southern blot (Fig.