Chapter 8: DNA: the Eukaryotic Chromosome Learning Objectives
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

(APOCI, -C2, and -E and LDLR) and the Genes C3, PEPD, and GPI (Whole-Arm Translocation/Somatic Cell Hybrids/Genomic Clones/Gene Family/Atherosclerosis) A
Proc. Natl. Acad. Sci. USA Vol. 83, pp. 3929-3933, June 1986 Genetics Regional mapping of human chromosome 19: Organization of genes for plasma lipid transport (APOCI, -C2, and -E and LDLR) and the genes C3, PEPD, and GPI (whole-arm translocation/somatic cell hybrids/genomic clones/gene family/atherosclerosis) A. J. LUSIS*t, C. HEINZMANN*, R. S. SPARKES*, J. SCOTTt, T. J. KNOTTt, R. GELLER§, M. C. SPARKES*, AND T. MOHANDAS§ *Departments of Medicine and Microbiology, University of California School of Medicine, Center for the Health Sciences, Los Angeles, CA 90024; tMolecular Medicine, Medical Research Council Clinical Research Centre, Harrow, Middlesex HA1 3UJ, United Kingdom; and §Department of Pediatrics, Harbor Medical Center, Torrance, CA 90509 Communicated by Richard E. Dickerson, February 6, 1986 ABSTRACT We report the regional mapping of human from defects in the expression of the low density lipoprotein chromosome 19 genes for three apolipoproteins and a lipopro- (LDL) receptor and is strongly correlated with atheroscle- tein receptor as well as genes for three other markers. The rosis (15). Another relatively common dyslipoproteinemia, regional mapping was made possible by the use of a reciprocal type III hyperlipoproteinemia, is associated with a structural whole-arm translocation between the long arm of chromosome variation of apolipoprotein E (apoE) (16). Also, a variety of 19 and the short arm of chromosome 1. Examination of three rare apolipoprotein deficiencies result in gross perturbations separate somatic cell hybrids containing the long arm but not of plasma lipid transport; for example, apoCII deficiency the short arm of chromosome 19 indicated that the genes for results in high fasting levels oftriacylglycerol (17). -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

GENOME GENERATION Glossary
GENOME GENERATION Glossary Chromosome An organism’s DNA is packaged into chromosomes. Humans have 23 pairs of chromosomesincluding one pair of sex chromosomes. Women have two X chromosomes and men have one X and one Y chromosome. Dominant (see also recessive) Genes come in pairs. A dominant form of a gene is the “stronger” version that will be expressed. Therefore if someone has one dominant and one recessive form of a gene, only the characteristics of the dominant form will appear. DNA DNA is the long molecule that contains the genetic instructions for nearly all living things. Two strands of DNA are twisted together into a double helix. The DNA code is made up of four chemical letters (A, C, G and T) which are commonly referred to as bases or nucleotides. Gene A gene is a section of DNA that is the code for a specific biological component, usually a protein. Each gene may have several alternative forms. Each of us has two copies of most of our genes, one copy inherited from each parent. Most of our traits are the result of the combined effects of a number of different genes. Very few traits are the result of just one gene. Genetic sequence The precise order of letters (bases) in a section of DNA. Genome A genome is the complete DNA instructions for an organism. The human genome contains 3 billion DNA letters and approximately 23,000 genes. Genomics Genomics is the study of genomes. This includes not only the DNA sequence itself, but also an understanding of the function and regulation of genes both individually and in combination. -

An Overview of the Independent Histories of the Human Y Chromosome and the Human Mitochondrial Chromosome
The Proceedings of the International Conference on Creationism Volume 8 Print Reference: Pages 133-151 Article 7 2018 An Overview of the Independent Histories of the Human Y Chromosome and the Human Mitochondrial chromosome Robert W. Carter Stephen Lee University of Idaho John C. Sanford Cornell University, Cornell University College of Agriculture and Life Sciences School of Integrative Plant Science,Follow this Plant and Biology additional Section works at: https://digitalcommons.cedarville.edu/icc_proceedings DigitalCommons@Cedarville provides a publication platform for fully open access journals, which means that all articles are available on the Internet to all users immediately upon publication. However, the opinions and sentiments expressed by the authors of articles published in our journals do not necessarily indicate the endorsement or reflect the views of DigitalCommons@Cedarville, the Centennial Library, or Cedarville University and its employees. The authors are solely responsible for the content of their work. Please address questions to [email protected]. Browse the contents of this volume of The Proceedings of the International Conference on Creationism. Recommended Citation Carter, R.W., S.S. Lee, and J.C. Sanford. An overview of the independent histories of the human Y- chromosome and the human mitochondrial chromosome. 2018. In Proceedings of the Eighth International Conference on Creationism, ed. J.H. Whitmore, pp. 133–151. Pittsburgh, Pennsylvania: Creation Science Fellowship. Carter, R.W., S.S. Lee, and J.C. Sanford. An overview of the independent histories of the human Y-chromosome and the human mitochondrial chromosome. 2018. In Proceedings of the Eighth International Conference on Creationism, ed. J.H. -

Cell Growth and Reproduction Lesson 6.2: Chromosomes and DNA Replication
Chapter 6: Cell Growth and Reproduction Lesson 6.2: Chromosomes and DNA Replication Cell reproduction involves a series of steps that always begin with the processes of interphase. During interphase the cell’s genetic information which is stored in its nucleus in the form of chromatin, composed of both mitotic and interphase chromosomes molecules of protein complexes and DNA strands that are loosely coiled winds tightly to be replicated. It is estimated that the DNA in human cells consists of approximately three billion nucleotides. If a DNA molecule was stretched out it would measure over 20 miles in length and all of it is stored in the microscopic nuclei of human cells. This lesson will help you to understand how such an enormous amount of DNA is coiled and packed in a complicated yet organized manner. During cell reproduction as a cell gets ready to divide the DNA coils even more into tightly compact structures. Lesson Objectives • Describe the coiled structure of chromosomes. • Understand that chromosomes are coiled structures made of DNA and proteins. They form after DNA replicates and are the form in which the genetic material goes through cell division. • Discover that DNA replication is semi-conservative; half of the parent DNA molecule is conserved in each of the two daughter DNA molecules. • Outline discoveries that led to knowledge of DNA’s structure and function. • Examine the processes of DNA replication. Vocabulary • centromere • double helix • Chargaff’s rules • histones • chromatid • nucleosomes • chromatin • semi-conservative DNA replication • chromosome • sister chromatids • DNA replication • transformation Introduction In eukaryotic cells, the nucleus divides before the cell itself divides. -

Metabolism As Related to Chromosome Structure and the Duration of Life by John W
METABOLISM AS RELATED TO CHROMOSOME STRUCTURE AND THE DURATION OF LIFE BY JOHN W. GOWEN (From the Department of Animal Pathology of The Rockefeller Institute for Medical Research, Princeton, N. ].) (Received for publication, December 16, 1930) In this paper it is proposed to measure the katabollsm of four fundamentally distinct groups of animals all within the same species and having closely similar genetic constitutions. These groups differ in what are perhaps the most significant elements of life. The chromosome structure of the first group is that of the type female, diploid; the second group is that of the type male, diploid but having one X and Y instead of the two X-chromosomes of the type female; the third group of flies are triploid, three sex chromosomes and three sets of autosomes; the fourth group, sex-intergrades has two X-chromo- somes and three sets of autosomes. Katabolism is a direct function of the cells composing the bodies of all animals. The cells of these four groups differ in size; the type male cells are the smallest; the females are somewhat, possibly a tenth, larger; the sex-intergrades and triploid cells are a half larger. The larger cell size suggests that any function within the cell would be performed in a larger way. The carbon dioxide production should enable us to measure physiologically the extent of this activity and present us with data on that fundamental point the relation of cell size to metabolic activity. The durations of life of these different groups have been measured. The forms with the balanced chromosome complexes within their cells live the longest time, the unbalanced groups the least. -

Genetic Variation in Chromosome Y Regulates Susceptibility to Influenza a Virus Infection
Genetic variation in chromosome Y regulates susceptibility to influenza A virus infection Dimitry N. Krementsova, Laure K. Casea, Oliver Dienzb, Abbas Razaa, Qian Fanga, Jennifer L. Athera, Matthew E. Poyntera, Jonathan E. Boysonb, Janice Y. Bunnc, and Cory Teuschera,d,1 aDepartment of Medicine, University of Vermont, Burlington, VT 05405; bDepartment of Surgery, University of Vermont, Burlington, VT 05405; cDepartment of Medical Biostatistics, University of Vermont, Burlington, VT 05405; and dDepartment of Pathology, University of Vermont, Burlington, VT 05405 Edited by Sabra Klein, Johns Hopkins University, Baltimore, MD, and accepted by Editorial Board Member Peter Palese January 23, 2017 (received for review December 20, 2016) Males of many species, ranging from humans to insects, are more encephalomyelitis (EAE) in SJL/J mice, which are controlled by susceptible than females to parasitic, fungal, bacterial, and viral genetic variation in ChrY (23–25). Moreover, with respect to infections. One mechanism that has been proposed to account for viral infections, we reported that genetic variation in ChrY this difference is the immunocompetence handicap model, which influences both survival following infection with Coxsackie- posits that the greater infectious disease burden in males is due to virus B3 virus (CVB3) (26) and susceptibility to CVB3-induced testosterone, which drives the development of secondary male sex autoimmune myocarditis (27). To address whether genetic vari- characteristics at the expense of suppressing immunity. However, ation in ChrY is capable of influencing susceptibility to IAV and emerging data suggest that cell-intrinsic (chromosome X and Y) the associated sex differences, we studied the susceptibility of a sex-specific factors also may contribute to the sex differences in panel of ChrY consomic strains on the C57BL/6J background infectious disease burden. -

Genesis of Non-Coding RNA Genes in Human Chromosome 22—A Sequence Connection with Protein Genes Separated by Evolutionary Time
non-coding RNA Perspective Genesis of Non-Coding RNA Genes in Human Chromosome 22—A Sequence Connection with Protein Genes Separated by Evolutionary Time Nicholas Delihas Department of Microbiology and Immunology, Renaissance School of Medicine, Stony Brook University, Stony Brook, New York, NY 11794-5222, USA; [email protected] Received: 16 July 2020; Accepted: 1 September 2020; Published: 3 September 2020 Abstract: A small phylogenetically conserved sequence of 11,231 bp, termed FAM247, is repeated in human chromosome 22 by segmental duplications. This sequence forms part of diverse genes that span evolutionary time, the protein genes being the earliest as they are present in zebrafish and/or mice genomes, and the long noncoding RNA genes and pseudogenes the most recent as they appear to be present only in the human genome. We propose that the conserved sequence provides a nucleation site for new gene development at evolutionarily conserved chromosomal loci where the FAM247 sequences reside. The FAM247 sequence also carries information in its open reading frames that provides protein exon amino acid sequences; one exon plays an integral role in immune system regulation, specifically, the function of ubiquitin-specific protease (USP18) in the regulation of interferon. An analysis of this multifaceted sequence and the genesis of genes that contain it is presented. Keywords: de novo gene birth; gene evolution; protogene; long noncoding RNA genes; pseudogenes; USP18; GGT5; Alu sequences 1. Introduction The genesis of genes has been a major topic of interest for several decades [1,2]. One mechanism of gene formation is by duplication of existing genes [1,3]. -

Basic Genetic Concepts & Terms
Basic Genetic Concepts & Terms 1 Genetics: what is it? t• Wha is genetics? – “Genetics is the study of heredity, the process in which a parent passes certain genes onto their children.” (http://www.nlm.nih.gov/medlineplus/ency/article/002048. htm) t• Wha does that mean? – Children inherit their biological parents’ genes that express specific traits, such as some physical characteristics, natural talents, and genetic disorders. 2 Word Match Activity Match the genetic terms to their corresponding parts of the illustration. • base pair • cell • chromosome • DNA (Deoxyribonucleic Acid) • double helix* • genes • nucleus Illustration Source: Talking Glossary of Genetic Terms http://www.genome.gov/ glossary/ 3 Word Match Activity • base pair • cell • chromosome • DNA (Deoxyribonucleic Acid) • double helix* • genes • nucleus Illustration Source: Talking Glossary of Genetic Terms http://www.genome.gov/ glossary/ 4 Genetic Concepts • H describes how some traits are passed from parents to their children. • The traits are expressed by g , which are small sections of DNA that are coded for specific traits. • Genes are found on ch . • Humans have two sets of (hint: a number) chromosomes—one set from each parent. 5 Genetic Concepts • Heredity describes how some traits are passed from parents to their children. • The traits are expressed by genes, which are small sections of DNA that are coded for specific traits. • Genes are found on chromosomes. • Humans have two sets of 23 chromosomes— one set from each parent. 6 Genetic Terms Use library resources to define the following words and write their definitions using your own words. – allele: – genes: – dominant : – recessive: – homozygous: – heterozygous: – genotype: – phenotype: – Mendelian Inheritance: 7 Mendelian Inheritance • The inherited traits are determined by genes that are passed from parents to children. -

Glossary/Index
Glossary 03/08/2004 9:58 AM Page 119 GLOSSARY/INDEX The numbers after each term represent the chapter in which it first appears. additive 2 allele 2 When an allele’s contribution to the variation in a One of two or more alternative forms of a gene; a single phenotype is separately measurable; the independent allele for each gene is inherited separately from each effects of alleles “add up.” Antonym of nonadditive. parent. ADHD/ADD 6 Alzheimer’s disease 5 Attention Deficit Hyperactivity Disorder/Attention A medical disorder causing the loss of memory, rea- Deficit Disorder. Neurobehavioral disorders character- soning, and language abilities. Protein residues called ized by an attention span or ability to concentrate that is plaques and tangles build up and interfere with brain less than expected for a person's age. With ADHD, there function. This disorder usually first appears in persons also is age-inappropriate hyperactivity, impulsive over age sixty-five. Compare to early-onset Alzheimer’s. behavior or lack of inhibition. There are several types of ADHD: a predominantly inattentive subtype, a predomi- amino acids 2 nantly hyperactive-impulsive subtype, and a combined Molecules that are combined to form proteins. subtype. The condition can be cognitive alone or both The sequence of amino acids in a protein, and hence pro- cognitive and behavioral. tein function, is determined by the genetic code. adoption study 4 amnesia 5 A type of research focused on families that include one Loss of memory, temporary or permanent, that can result or more children raised by persons other than their from brain injury, illness, or trauma. -
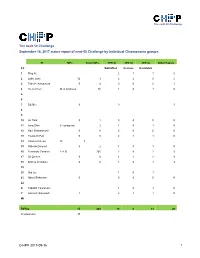
Next-MP50 Status Report
The neXt-50 Challenge The neXt-50 Challenge September 16, 2017 status report of next-50 Challenge by individual Chromosome groups. PI MPs Silver MPs JPR SI JPR SI JPR SI Other Papers Ch Submitted In press In revision 1 Ping Xu 2 1 1 0 2 Lydie Lane 12 3 2 2 0 2 3 Takeshi Kawamura 0 4 0 0 0 1 4 Yu-Ju Chen 26 in progress 86 1 0 1 0 5 6 7 Ed Nice 0 0 1 8 9 10 Jin Park 0 1 0 0 0 0 11 Jong Shin 2 in progress 2 1 0 1 0 12 Ravi Sirdeshmukh 0 0 0 0 0 0 13 Young-Ki Paik 0 5 2 1 1 0 14 CharLes Pineau 12 3 15 GiLberto Domont 0 2 1 0 1 0 16 Fernando CorraLes 2 (+ 9) 165 2 0 2 5 17 GiL Omenn 0 0 3 1 2 8 18 Andrey Archakov 0 0 1 0 1 3 19 20 Siqi Liu 1 0 1 21 Albert Sickmann 0 0 0 0 0 22 X Tadashi Yamamoto 1 0 1 0 Y Hosseini SaLekdeh 1 2 1 1 0 Mt TOTAL 15 268 19 6 13 20 (in progress) 37 C-HPP 2017-09-16 1 The neXt-50 Challenge Chromosome 1 (Ping Xu) PIC Leaders: Ping Xu, Fuchu He Contributing labs: Ping Xu, Beijing Proteome Research Center Fuchu He, Beijing Proteome Research Center Dong Yang, Beijing Proteome Research Center Wantao Ying, Beijing Proteome Research Center Pengyuan Yang, Fudan University Siqi Liu, Beijing Genome Institute Qinyu He, Jinan University Major lab members or partners contributing to the neXt50: Yao Zhang (Beijing Proteome Research Center), Yihao Wang (Beijing Proteome Research Center), Cuitong He (Beijing Proteome Research Center), Wei Wei (Beijing Proteome Research Center), Yanchang Li (Beijing Proteome Research Center), Feng Xu (Beijing Proteome Research Center), Xuehui Peng (Beijing Proteome Research Center). -

Initiating Chromosome Replication in E. Coli: It Makes Sense to Recycle
Downloaded from genesdev.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press PERSPECTIVE Initiating chromosome replication in E. coli: it makes sense to recycle Alan C. Leonard1 and Julia E. Grimwade2 Department of Biological Sciences, Florida Institute of Technology, Melbourne, Florida 32901, USA Initiating new rounds of Escherichia coli chromosome Escherichia coli initiator DnaA: building a bacterial replication requires DnaA-ATP to unwind the replication ORC and pre-RC origin, oriC, and load DNA helicase. In this issue of Questions about cycling initiator activity are particularly Genes & Development, Fujimitsu and colleagues (pp. relevant to DNA replication control in rapidly growing 1221–1233) demonstrate that two chromosomal sites, bacteria, since these cells contain multiple origins that termed DARS (DnaA-reactivating sequences), recycle must fire synchronously only once per cell division cycle inactive DnaA-ADP into DnaA-ATP. Fujimitsu and col- (Skarstad et al. 1986; Boye et al. 2000). Studies of the E. leagues propose these sites are necessary to attain the coli AAA+ initiator, DnaA protein, provide insights into DnaA-ATP threshold during normal growth and are im- portant regulators of initiation timing in bacteria. the various cellular mechanisms that ensure that the initiator is active and available at the time of initiation and is then prevented from triggering another round of replication until the next cell cycle. DnaA has a high All cells must coordinate the timing of chromosome affinity for adenine nucleotides ADP and ATP, but is duplication with cell division during the cell cycle, to active only when bound to ATP (Sekimizu et al.