Comprehensive Review of 3D Segmentation Software Tools for MRI Usable for Pelvic Surgery Planning
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

Modern Programming Languages CS508 Virtual University of Pakistan
Modern Programming Languages (CS508) VU Modern Programming Languages CS508 Virtual University of Pakistan Leaders in Education Technology 1 © Copyright Virtual University of Pakistan Modern Programming Languages (CS508) VU TABLE of CONTENTS Course Objectives...........................................................................................................................4 Introduction and Historical Background (Lecture 1-8)..............................................................5 Language Evaluation Criterion.....................................................................................................6 Language Evaluation Criterion...................................................................................................15 An Introduction to SNOBOL (Lecture 9-12).............................................................................32 Ada Programming Language: An Introduction (Lecture 13-17).............................................45 LISP Programming Language: An Introduction (Lecture 18-21)...........................................63 PROLOG - Programming in Logic (Lecture 22-26) .................................................................77 Java Programming Language (Lecture 27-30)..........................................................................92 C# Programming Language (Lecture 31-34) ...........................................................................111 PHP – Personal Home Page PHP: Hypertext Preprocessor (Lecture 35-37)........................129 Modern Programming Languages-JavaScript -

A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours
A 3D interactive multi-object segmentation tool using local robust statistics driven active contours The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Gao, Yi, Ron Kikinis, Sylvain Bouix, Martha Shenton, and Allen Tannenbaum. 2012. A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours. Medical Image Analysis 16, no. 6: 1216–1227. doi:10.1016/j.media.2012.06.002. Published Version doi:10.1016/j.media.2012.06.002 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:28548930 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA NIH Public Access Author Manuscript Med Image Anal. Author manuscript; available in PMC 2013 August 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Med Image Anal. 2012 August ; 16(6): 1216–1227. doi:10.1016/j.media.2012.06.002. A 3D Interactive Multi-object Segmentation Tool using Local Robust Statistics Driven Active Contours Yi Gaoa,*, Ron Kikinisb, Sylvain Bouixa, Martha Shentona, and Allen Tannenbaumc aPsychiatry Neuroimaging Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 bSurgical Planning Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 cDepartments of Electrical and Computer Engineering and Biomedical Engineering, Boston University, Boston, MA 02115 Abstract Extracting anatomical and functional significant structures renders one of the important tasks for both the theoretical study of the medical image analysis, and the clinical and practical community. -

TANGO Device
School on TANGO Controls system Basics of TANGO Lorenzo Pivetta Claudio Scafuri Graziano Scalamera http://www.tango-controls.org L.Pivetta, C.Scafuri, G.Scalamera School on TANGO Control System - Trieste 4-8th July 2016 2 Prerequisites To better understand the training a background on the following arguments is desirable: ● Programming language ● Object oriented programming ● Linux/UNIX operating system ● Networking ● Control systems L.Pivetta, C.Scafuri, G.Scalamera School on TANGO Control System - Trieste 4-8th July 2016 3 Outline 1 - What is TANGO? 2 - TANGO architecture Language/OS/Compilers Device hierarchy CORBA and ZeroMQ TANGO domains TANGO device and device server TANGO Database Communication models 3 - TANGO configuration/tools Multicast Jive Polling Starter/Astor Events Pogo Alarms TANGO installation Groups Client basics TANGO ACL Logging system Historical DataBase 4 – Examples Test device L.Pivetta, C.Scafuri, G.Scalamera School on TANGO Control System - Trieste 4-8th July 2016 4 What is TANGO? Scientific workspaces Native client applications In short: Industrial SCADA Control system framework TANGO Based on CORBA and ZMQ C++ TANGO Java Archiving Python System Centralized config. database TANGO TANGO TANGO TANGO Clients binding binding binding binding (CLI/GUI) Software bus for distributed TANGO software bus objects Provides unified interface to Device Device Device Device Device Device Device Server Server Server Server Server Server Server all equipments hiding how they are HV ps + Pylon OPC UA Data Motion TANGO SNMP connected/managed -

Accurate Segmentation of Brain MR Images
Accurate segmentation of brain MR images Master of Science Thesis in Biomedical Engineering ANTONIO REYES PORRAS PÉREZ Department of Signals and Systems Division of Biomedical Engineering CHALMERS UNIVERSITY OF TECHNOLOGY Göteborg, Sweden, 2010 Report No. EX028/2010 Abstract Full brain segmentation has been of significant interest throughout the years. Recently, many research groups worldwide have been looking into development of patient-specific electromagnetic models for dipole source location in EEG. To obtain this model, accurate segmentation of various tissues and sub-cortical structures is thus required. In this project, the performance of three of the most widely used software packages for brain segmentation has been analyzed: FSL, SPM and FreeSurfer. For the analysis, real images from a patient and a set of phantom images have been used in order to evaluate the performance r of each one of these tools. Keywords: dipole source location, brain, patient-specific model, image segmentation, FSL, SPM, FreeSurfer. Acknowledgements To my advisor, Antony, for his guidance through the project. To my partner, Koushyar, for all the days we have spent in the hospital helping each other. To the staff in Sahlgrenska hospital for their collaboration. To MedTech West for this opportunity to learn. Table of contents 1. Introduction ......................................................................................................................................... 1 2. Magnetic resonance imaging .............................................................................................................. -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

A Practical Guide for Improving Transparency and Reproducibility in Neuroimaging Research Krzysztof J
bioRxiv preprint first posted online Feb. 12, 2016; doi: http://dx.doi.org/10.1101/039354. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. It is made available under a CC-BY 4.0 International license. A practical guide for improving transparency and reproducibility in neuroimaging research Krzysztof J. Gorgolewski and Russell A. Poldrack Department of Psychology, Stanford University Abstract Recent years have seen an increase in alarming signals regarding the lack of replicability in neuroscience, psychology, and other related fields. To avoid a widespread crisis in neuroimaging research and consequent loss of credibility in the public eye, we need to improve how we do science. This article aims to be a practical guide for researchers at any stage of their careers that will help them make their research more reproducible and transparent while minimizing the additional effort that this might require. The guide covers three major topics in open science (data, code, and publications) and offers practical advice as well as highlighting advantages of adopting more open research practices that go beyond improved transparency and reproducibility. Introduction The question of how the brain creates the mind has captivated humankind for thousands of years. With recent advances in human in vivo brain imaging, we how have effective tools to peek into biological underpinnings of mind and behavior. Even though we are no longer constrained just to philosophical thought experiments and behavioral observations (which undoubtedly are extremely useful), the question at hand has not gotten any easier. These powerful new tools have largely demonstrated just how complex the biological bases of behavior actually are. -

1 Structural and Functional Alterations in the Brain Gray Matter
Structural and functional alterations in the brain gray matter among first-degree relatives of schizophrenia patients: a multimodal meta-analysis of fMRI and VBM studies Running title: Familial risk for schizophrenia and alterations in the brain Aino I. L. Saarinen, PhD1,2,3,*, Sanna Huhtaniska, MD, PhD4, Juho Pudas, MB3, Lassi Björnholm, MD3, Tuomas Jukuri, MD, PhD3, Jussi Tohka, MD, PhD5, Niklas Granö, PhD6, Jennifer H. Barnett, PhD7,8, Vesa Kiviniemi, MD, PhD9,10, Juha Veijola, MD, PhD3,10,11, Mirka Hintsanen, PhD1, Johannes Lieslehto, MD, PhD 12 1 Research Unit of Psychology, University of Oulu, Finland 2 Department of Psychology and Logopedics, Faculty of Medicine, University of Helsinki, Finland 3 Research Unit of Clinical Neuroscience, Department of Psychiatry, University of Oulu, Finland 4 Center for Life Course Health Research, University of Oulu, Finland 5 A.I. Virtanen Institute for Molecular Sciences, University of Eastern Finland, Kuopio, Finland 6 Helsinki University Hospital, Department of Adolescent Psychiatry, Finland 7 Cambridge Cognition, Cambridge, UK 8 Department of Psychiatry, University of Cambridge, Cambridge, UK 9 Department of Diagnostic Radiology, Oulu University Hospital, Oulu, Finland 10 Department of Psychiatry, Oulu University Hospital, Oulu, Finland 11 Medical Research Center Oulu, Oulu University Hospital and University of Oulu, Oulu, Finland 12 Section for Neurodiagnostic Applications, Department of Psychiatry, Ludwig Maximilian University, Nussbaumstrasse 7, 80336 Munich, Bavaria, * Corresponding author: Aino Saarinen. Department of Psychology and Logopedics, Faculty of Medicine, Haartmaninkatu 3, P.O. Box 21, 00014 University of Helsinki, Finland. E-mail: [email protected], Tel.: +35844 307 1204. 1 Abstract Objective: Schizophrenia has one of the highest heritability estimates in psychiatry, but the genetically- based underlying neuropathology has mainly remained unclear. -

Simultaneous PET/MRI for Connectivity Mapping: Quantitative Methods in Clinical Setting
UNIVERSITY OF PADOVA Department of Information Engineering Ph.D. School on Information Engineering Curriculum: Bioengineering - Cycle: XXX Simultaneous PET/MRI for Connectivity Mapping: Quantitative Methods in Clinical Setting Headmaster of the School: Prof. Andrea NEVIANI Supervisor: Prof. Alessandra BERTOLDO PhD. Candidate: Eng. Erica SILVESTRI 2018 iii University of Padova Abstract Ph.D. School on Information Engineering Simultaneous PET/MRI for Connectivity Mapping: Quantitative Methods in Clinical Setting by Eng. Erica SILVESTRI In recent years, the study of brain connectivity has received growing inter- est from neuroscience field, from a point of view both of analysis of patho- logical condition and of a healthy brain. Hybrid PET/MRI scanners are promising tools to study this complex phenomenon. This thesis presents a general framework for the acquisition and analysis of simultaneous multi- modal PET/MRI imaging data to study brain connectivity in a clinical set- ting. Several aspects are faced ranging from the planning of an acquisition protocol consistent with clinical constraint to the off-line PET image recon- struction, from the selection and implementation of methods for quantifying the acquired data to the development of methodologies to combine the com- plementary informations obtained with the two modalities. The developed analysis framework was applied to two different studies, a first conducted on patients affected by Parkinson’s Disease and dementia, and a second one on high grade gliomas, as proof of concept evaluation that the pipeline can be extended in clinical settings. v Università degli Studi di Padova Sommario Scuola di Dottorato in Ingegneria dell’Informazione Acquisizioni simultanee PET/MR per lo studio della connettività: metodi quantitativi in ambito clinico di Ing. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -
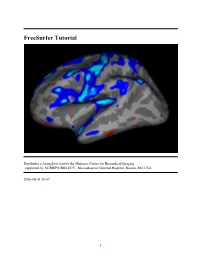
Freesurfer Tutorial
FreeSurfer Tutorial FreeSurfer is brought to you by the Martinos Center for Biomedical Imaging supported by NCRR/P41RR14075, Massachusetts General Hospital, Boston, MA USA 2008-06-01 20:47 1 FreeSurfer Tutorial Table of Contents Section Page Overview and course outline 3 Inspection of Freesurfer output 5 Troubleshooting your output 22 Fixing a bad skull strip 26 Making edits to the white matter 34 Correcting pial surfaces 46 Using control points to fix intensity normalization 50 Talairach registration 55 recon-all: morphometry and reconstruction 69 recon-all: process flow table 97 QDEC Group analysis 100 Group analysis: average subject, design matrix, mri_glmfit 125 Group analysis: visualization and inspection 149 Integrating FreeSurfer and FSL's FEAT 159 Exercise overview 172 Tkmedit reference 176 Tksurfer reference 211 Glossary 227 References 229 Acknowledgments 238 2008-06-01 20:47 2 top FreeSurfer Slides 1. Introduction to Freesurfer - Bruce Fischl 2. Anatomical Analysis with Freesurfer - Doug Greve 3. Surface-based Group Analysis - Doug Greve 4. Applying FreeSurfer Tools to FSL fMRI Analysis - Doug Greve FreeSurfer Tutorial Overview The FreeSurfer tools deal with two main types of data: volumetric data volumes of voxels and surface data polygons that tile a surface. This tutorial should familiarize you with FreeSurfer’s volume and surface processing streams, the recommended workflow to execute these, and many of their component tools. The tutorial also describes some of FreeSurfer's tools for registering volumetric datasets, performing group analysis on morphology data, and integrating FSL Feat output with FreeSurfer overlaying color coded parametric maps onto the cortical surface and visualizing plotted results. -

Confrontsred Tape
IPS: THE MENTAL HEALTH SERVICES CONFERENCE Oct. 8-11, 2015 • New York City When Good Care Confronts Red Tape Navigating the System for Our Patients and Our Practice New York, NY | Sheraton New York Times Square POSTER SESSION OCTOBER 09, 2015 POSTER SESSION 1 P1- 1 ANGIOEDEMA DUE TO POTENTIATING EFFECTS OF RITONIVIR ON RISPERIDONE: CASE REPORT Lead Author: Luisa S. Gonzalez, M.D. Co-Author(s): David A. Kasle MSIII Kavita Kothari M.D. SUMMARY: For many drugs, the liver is the principal site of its metabolism. The most important enzyme system of phase I metabolism is the cytochrome P-450 (CYP450). Ritonavir, a protease inhibitor used for the treatment of human immunodeficiency virus (HIV) infection and acquired immunodeficiency syndrome (AIDS), has been shown to be a potent inhibitor of the (CYP450) 3A and 2D6 isozymes. This inhibition increases the chance for potential drug-drug interactions with compounds that are metabolized by these isoforms. Risperidone, a second generation atypical antipsychotic, is metabolized to a significant extent by the (CYP450) 2D6, and to a lesser extent 3A4. Ritonavir and risperidone can both cause serious, and even life threatening, side effects. An example of one such rare side effect is angioedema which is characterized by edema of the deep dermal and subcutaneous tissues. Here we present three cases of patients residing in a nursing home setting, diagnosed with HIV/AIDS and comorbid psychiatric illness. These patients were all on antiretroviral medications such as ritonavir, and all later presented with varying degrees of edema after long term use of risperidone. We aim to bring to awareness the potential for ritonavir inhibiting the metabolism of risperidone, thus leading to an increased incidence of angioedema in these patients.