Post-Transcriptional Control of EMT Is Coordinated Through Combinatorial
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Investigation of Adiposity Phenotypes in AA Associated with GALNT10 & Related Pathway Genes
Investigation of Adiposity Phenotypes in AA Associated With GALNT10 & Related Pathway Genes By Mary E. Stromberg A Dissertation Submitted to the Graduate Faculty of WAKE FOREST UNIVERSITY GRADUATE SCHOOL OF ARTS AND SCIENCES in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY In Molecular Genetics and Genomics December 2018 Winston-Salem, North Carolina Approved by: Donald W. Bowden, Ph.D., Advisor Maggie C.Y. Ng, Ph.D., Advisor Timothy D. Howard, Ph.D., Chair Swapan Das, Ph.D. John P. Parks, Ph.D. Acknowledgements I would first like to thank my mentors, Dr. Bowden and Dr. Ng, for guiding my learning and growth during my years at Wake Forest University School of Medicine. Thank you Dr. Ng for spending so much time ensuring that I learn every detail of every protocol, and supporting me through personal difficulties over the years. Thank you Dr. Bowden for your guidance in making me a better scientist and person. I would like to thank my committee for their patience and the countless meetings we have had in discussing this project. I would like to say thank you to the members of our lab as well as the Parks lab for their support and friendship as well as their contributions to my project. Special thanks to Dean Godwin for his support and understanding. The umbrella program here at WFU has given me the chance to meet some of the best friends I could have wished for. I would like to also thank those who have taught me along the way and helped me to get to this point of my life, with special thanks to the late Dr. -

Identification, Characterization, and Utilization of Glycosyltransferases
Identification, Characterization, and Utilization of Glycosyltransferases DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Nicholas Roman Pettit Graduate Program in Chemistry The Ohio State University 2011 Dissertation Committee: Dr. Peng George Wang, Advisor Dr. Zucai Suo Dr. Karin Musier-Forsyth Copyright by Nicholas Pettit 2011 3 ABSTRACT Among the many biomolecules found in nature, such as amino acids and DNA, carbohydrates are among the most important and abundant. The assembly of carbohydrates to build various oligosaccharides or glycans is of considerable interest in the glycobiology community, as understanding, controlling, and manipulating such assemblies can yield many interesting insights. This is because glycans are directly involved with many critical biological processes such as molecular recognition, cell-to- cell communication, protein folding and stability, as well as being involved many unique disease state phenotypes. The assembly of these glycans is commonly accomplished by a class of enzymes called glycosyltransferases, and it is these enzymes which have been exploited in vitro for the synthesis of attractive oligosaccharides. However, the identification, characterization, and subsequent exploitation of a glycosyltransferase is a very tedious and long process which stems from the root problem that knowing the primary function of a glycosyltransferase must be determined experimentally. This is because glycans -

Molecular Evolution of the Glycosyltransferase 6 Gene Family in Primates
Hindawi Publishing Corporation Biochemistry Research International Volume 2016, Article ID 9051727, 6 pages http://dx.doi.org/10.1155/2016/9051727 Research Article Molecular Evolution of the Glycosyltransferase 6 Gene Family in Primates Eliane Evanovich,1 Patricia Jeanne de Souza Mendonça-Mattos,1 and Maria Lúcia Harada2 1 Laboratorio´ de Genetica´ Humana e Medica,´ Instituto de Cienciasˆ Biologicas,´ Universidade Federal do Para,´ Belem,´ PA, Brazil 2Laboratorio´ de Biologia Molecular “Francisco Mauro Salzano”, Instituto de Cienciasˆ Biologicas,´ Universidade Federal do Para,´ Belem,´ PA, Brazil Correspondence should be addressed to Eliane Evanovich; [email protected] Received 12 July 2016; Accepted 20 October 2016 Academic Editor: Stefano Pascarella Copyright © 2016 Eliane Evanovich et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Glycosyltransferase 6 gene family includes ABO, Ggta1, iGb3S, and GBGT1 genes and by three putative genes restricted to mammals, GT6m6, GTm6, and GT6m7, only the latter is found in primates. GT6 genes may encode functional and nonfunctional proteins. Ggta1 and GBGT1 genes, for instance, are pseudogenes in catarrhine primates, while iGb3S gene is only inactive in human, bonobo, and chimpanzee. Even inactivated, these genes tend to be conversed in primates. As some of the GT6 genes are related to the susceptibility or resistance to parasites, we investigated (i) the selective pressure on the GT6 paralogs genes in primates; (ii) the basis of the conservation of iGb3S in human, chimpanzee, and bonobo; and (iii) the functional potential of the GBGT1 and GT6m7 in catarrhines. -

Human Social Genomics in the Multi-Ethnic Study of Atherosclerosis
Getting “Under the Skin”: Human Social Genomics in the Multi-Ethnic Study of Atherosclerosis by Kristen Monét Brown A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Epidemiological Science) in the University of Michigan 2017 Doctoral Committee: Professor Ana V. Diez-Roux, Co-Chair, Drexel University Professor Sharon R. Kardia, Co-Chair Professor Bhramar Mukherjee Assistant Professor Belinda Needham Assistant Professor Jennifer A. Smith © Kristen Monét Brown, 2017 [email protected] ORCID iD: 0000-0002-9955-0568 Dedication I dedicate this dissertation to my grandmother, Gertrude Delores Hampton. Nanny, no one wanted to see me become “Dr. Brown” more than you. I know that you are standing over the bannister of heaven smiling and beaming with pride. I love you more than my words could ever fully express. ii Acknowledgements First, I give honor to God, who is the head of my life. Truly, without Him, none of this would be possible. Countless times throughout this doctoral journey I have relied my favorite scripture, “And we know that all things work together for good, to them that love God, to them who are called according to His purpose (Romans 8:28).” Secondly, I acknowledge my parents, James and Marilyn Brown. From an early age, you two instilled in me the value of education and have been my biggest cheerleaders throughout my entire life. I thank you for your unconditional love, encouragement, sacrifices, and support. I would not be here today without you. I truly thank God that out of the all of the people in the world that He could have chosen to be my parents, that He chose the two of you. -
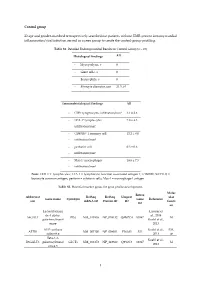
1 Control Group 23 Age and Gender-Matched
Control group 23 age and gender-matched retrospectively searched out patients without EMB-proven intramyocardial inflammation/viral infection served as a peer group to create the control-group profiling. Table S1. Detailed Endomyocardial Results in Control Group (n = 23) Histological findings All - Myocytolysis, n 0 - Giant cells, n 0 - Eosinophils, n 0 - Myocyte diameter, µm 21.3 ± 0 Immunohistological findings All - CD3+ lymphocytes infiltration/mm² 3.1 ± 2.6 - LFA-1+ lymphocytes 7.8 ± 4.3 infiltration/mm² - CD45R0+ T memory cell 12.2 ± 8.0 - infiltration/mm² - perforin+ cell 0.3 ± 0.6 - infiltration/mm² - Mac-1+ macrophages 18.6 ± 7.5 - infiltration/mm² Note: CD3 = T lymphocytes; LFA-1 = lymphocyte function-associated antigen-1; CD45R0 (UCHL1) = leucocyte common antigen; perforin = cytotoxic cells; Mac-1 = macrophage-1 antigen. Table S2. Potential marker genes for gene profile development. Molec Entrez Abbreviat RefSeq RefSeq Uniprot ular Gene name Synonym Gene Reference ion mRNA ID Protein ID ID functi ID on Lactosylcerami Lassner et de 4-alpha- al., 2014 A4GALT P(k) NM_017436 NP_059132 Q9NPC4 53947 M galactosyltransf Kuehl et al., erase 2013 ATP synthase Kuehl et al., EM, ATP5I NM_007100 NP_009031 P56385 521 subunit g 2013 ce Beta-1,3- Kuehl et al., B3GALT5 galactosyltransf GLCT5 NM_033172 NP_149362 Q9Y2C3 10317 M 2013 erase 5 1 BAG family molecular Kuehl et al., BAG5 NM_004873 NP_004864 Q9UL15 9529 CC chaperone 2013 regulator 5 Lassner et CC-chemokine al., 2014 CCL20 ST38 NM_004591 NP_004582 P78556 6364 IR ligand 20 Kuehl et al., -

A Novel 2.3 Mb Microduplication of 9Q34. 3 Inserted Into 19Q13. 4 in A
Hindawi Publishing Corporation Case Reports in Pediatrics Volume 2012, Article ID 459602, 7 pages doi:10.1155/2012/459602 Case Report A Novel 2.3 Mb Microduplication of 9q34.3 Inserted into 19q13.4 in a Patient with Learning Disabilities Shalinder Singh,1 Fern Ashton,1 Renate Marquis-Nicholson,1 Jennifer M. Love,1 Chuan-Ching Lan,1 Salim Aftimos,2 Alice M. George,1 and Donald R. Love1, 3 1 Diagnostic Genetics, LabPlus, Auckland City Hospital, P.O. Box 110031, Auckland 1148, New Zealand 2 Genetic Health Service New Zealand-Northern Hub, Auckland City Hospital, Private Bag 92024, Auckland 1142, New Zealand 3 School of Biological Sciences, University of Auckland, Private Bag 92019, Auckland 1142, New Zealand Correspondence should be addressed to Donald R. Love, [email protected] Received 1 July 2012; Accepted 27 September 2012 Academic Editors: L. Cvitanovic-Sojat, G. Singer, and V. C. Wong Copyright © 2012 Shalinder Singh et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Insertional translocations in which a duplicated region of one chromosome is inserted into another chromosome are very rare. We report a 16.5-year-old girl with a terminal duplication at 9q34.3 of paternal origin inserted into 19q13.4. Chromosomal analysis revealed the karyotype 46,XX,der(19)ins(19;9)(q13.4;q34.3q34.3)pat. Cytogenetic microarray analysis (CMA) identified a ∼2.3Mb duplication of 9q34.3 → qter, which was confirmed by Fluorescence in situ hybridisation (FISH). -

Supplementary Appendix Table of Contents
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Table of Contents Supplementary Note ........................................................................................................................... 1 Author contributions ............................................................................................................................... 1 Additional authors from study groups .............................................................................................. 2 Task Force COVID-19 Humanitas ......................................................................................................................................... 2 TASK FORCE COVID-19 HUMANITAS GAVAZZENI & CASTELLI ............................................................................. 3 COVICAT Study Group ............................................................................................................................................................... 4 Pa COVID-19 Study Group ....................................................................................................................................................... 5 Covid-19 Aachen Study (COVAS) .......................................................................................................................................... 5 Norwegian SARS-CoV-2 Study group ................................................................................................................................. -

Review of a Blood Group System Reborn
R EVIEW The FORS awakens: review of a blood group system reborn A.K. Hult and M.L. Olsson The presence of the FORS1 antigen on red blood cells was fetomaternal incompatibility. This review will summarize the discovered relatively recently, and in 2012, the International current knowledge on the newest of our carbohydrate blood Society of Blood Transfusion (ISBT) acknowledged FORS as group systems, FORS, acknowledged by the International blood group system number 031. This rare antigen is carried by a glycosphingolipid and formed by elongation of the P antigen. Society of Blood Transfusion (ISBT) in 2012. For obvious Most people have naturally occurring anti-FORS1 in their plasma. reasons, the body of knowledge is still limited, especially when The clinical significance of these antibodies is unknown in the it comes to the clinical significance of this system. transfusion setting, but they can hemolyze FORS1+ erythrocytes in the presence of complement in vitro. First believed to be part of the ABO system, it was later shown that the gene encoding the Forssman: Historical Aspects glycosyltransferase giving rise to FORS1 expression is GBGT1. This gene had previously been deemed nonfunctional in humans, The first mention of what was later designated the but a mutation, so far only detected in FORS1+ individuals, 9 restores the enzymatic activity. Tissue distribution of the antigen Forssman (Fs) antigen was reported in 1907 by Frouin. in FORS1+ individuals has not been studied in detail, although This heterophilic antigen was re-identified in 1911 by John the gene is expressed in several cell types. The antigen itself is Forssman (1868–1947), professor of microbiology, pathology, known to be a receptor for various pathogens and toxins and has and general medicine at Lund University, Lund, Sweden been detected in different forms of cancer, but the implications thereof are not fully understood. -

Supplementary Figure 1. Dystrophic Mice Show Unbalanced Stem Cell Niche
Supplementary Figure 1. Dystrophic mice show unbalanced stem cell niche. (A) Single channel images for the merged panels shown in Figure 1A, for of PAX7, MYOD and Laminin immunohistochemical staining in Lmna Δ8-11 mice of PAX7 and MYOD markers at the indicated days of post-natal growth. Basement membrane of muscle fibers was stained with Laminin. Scale bars, 50 µm. (B) Quantification of the % of PAX7+ MuSCs per 100 fibers at the indicated days of post-natal growth in (A). n =3-6 animals per genotype. (C) Immunohistochemical staining in Lmna Δ8-11 mice of activated, ASCs (PAX7+/KI67+) and quiescent QSCs (PAX7+/Ki67-) MuSCs at d19 and relative quantification (below). n= 4-6 animals per genotype. Scale bars, 50 µm. (D) Quantification of the number of cells per cluster in single myofibers extracted from d19 Lmna Δ8-11 mice and cultured 96h. n= 4-5 animals per group. Data are box with median and whiskers min to max. B, C, Data are mean ± s.e.m. Statistics by one-way (B) or two-way (C, D) analysis of variance (ANOVA) with multiple comparisons. * * P < 0.01, * * * P < 0.001. wt= Lmna Δ8-11 +/+; het= Lmna Δ8-11 +/; hom= Lmna Δ8-11 -/-. Supplementary Figure 2. Heterozygous mice show intermediate Lamin A levels. (A) RNA-seq signal tracks as the effective genome size normalized coverage of each biological replicate of Lmna Δ8-11 mice on Lmna locus. Neomycine cassette is indicated as a dark blue rectangle. (B) Western blot of total protein extracted from the whole Lmna Δ8-11 muscles at d19 hybridized with indicated antibodies. -

Molecular Genetics and Genomics of the ABO Blood Group System
19 Review Article Page 1 of 19 Molecular genetics and genomics of the ABO blood group system Fumiichiro Yamamoto Immunohematology and Glycobiology, Josep Carreras Leukaemia Research Institute (IJC), Badalona, Spain Correspondence to: Fumiichiro Yamamoto. Immunohematology and Glycobiology, Josep Carreras Leukaemia Research Institute (IJC), Badalona, Spain. Email: [email protected]. Abstract: The A and B oligosaccharide antigens of the ABO blood group system are produced from the common precursor, H substance, by enzymatic reactions catalyzed by A and B glycosyltransferases (AT and BT) encoded by functional A and B alleles at the ABO genetic locus, respectively. In 1990, my research team cloned human A, B, and O allelic cDNAs. We then demonstrated this central dogma of ABO and opened a new era of molecular genetics. We identified four amino acid substitutions between AT and BT and inactivating mutations in the O alleles, clarifying the allelic basis of ABO. We became the first to achieve successful ABO genotyping, discriminating between AA and AO genotypes and between BB and BO, which was impossible using immunohematological/serological methods. We also identified mutations in several subgroup alleles and also in the cis-AB and B(A) alleles that specify the expression of the A and B antigens by single alleles. Later, other scientists interested in the ABO system characterized many additional ABO alleles. However, the situation has changed drastically in the last decade, due to rapid advances in next- generation sequencing (NGS) technology, which has allowed the sequencing of several thousand genes and even the entire genome in individual experiments. Genome sequencing has revealed not only the exome but also transcription/translation regulatory elements.