Essential Role of GATA5 in the Mammalian Heart
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Embryology and Anatomy of Fetal Heart
Prof. Saeed Abuel Makarem Dr. Jamila El Medany Objectives • By the end of this lecture the student should be able to: • Describe the formation, sit, union divisions of the of the heart tubes. • Describe the formation and fate of the sinus venosus. • Describe the partitioning of the common atrium and common ventricle. • Describe the partitioning of the truncus arteriosus. • List the most common cardiac anomalies. • The CVS is the first major system to function in the embryo. • The heart begins to beat at (22nd – 23rd ) days. • Blood flow begins during the beginning of the fourth week and can be visualized by Ultrasound Doppler Notochord: stimulates neural tube formation Somatic mesoderm Splanchnic mesoderm FORMATION OF THE HEART TUBE • The heart is the first functional organ to develop. • It develops from Splanchnic Mesoderm in the wall of the yolk sac (Cardiogenic Area): Cranial to the developing Mouth & Nervous system and Ventral to the developing Pericardial sac. • The heart primordium is first evident at day 18 (as an Angioplastic cords which soon canalize to form the 2 heart tubes). • As the Head Fold completed, the developing heart tubes change their position and become in the Ventral aspect of the embryo, Dorsal to the developing Pericardial sac. • . Development of the Heart tube • After Lateral Folding of the embryo, the 2 heart tubes approach each other and fuse to form a single Endocardial Heart tube within the pericardial sac. • Fusion of the two tubes occurs in a Craniocaudal direction. What is the • The heart tube grows faster than shape of the the pericardial sac, so it shows 5 alternate dilations separated by Heart Tube? constrictions. -

Goals and Outcomes – Gametogenesis, Fertilization (Embryology Chapter 1)
Department of Histology and Embryology, Faculty of Medicine in Pilsen, Charles University, Czech Republic; License Creative Commons - http://creativecommons.org/licenses/by-nc-nd/3.0/ Goals and outcomes – Gametogenesis, fertilization (Embryology chapter 1) Be able to: − Define and use: progenesis, gametogenesis, primordial gonocytes, spermatogonia, primary and secondary spermatocytes, spermatids, sperm cells (spermatozoa), oogonia, primary and secondary oocytes, polar bodies, ovarian follicles (primordial, primary, secondary, tertiary), membrane granulosa, cumulus oophorus, follicular antrum, theca folliculi interna and externa, zona pellucida, corona radiata, ovulation, corpus luteum, corpus albicans, follicular atresia, expanded cumulus, luteinizing hormone (LH), follicle-stimulating hormone (FSH), human chorionic gonadotropin (hCG), sperm capacitation, acrosome reaction, cortical reaction and zona reaction, fertilization, zygote, cleavage, implantation, gastrulation, organogenesis, embryo, fetus, cell division, differentiation, morphogenesis, condensation, migration, delamination, apoptosis, induction, genotype, phenotype, epigenetics, ART – assisted reproductive techniques, spermiogram, IVF-ET (in vitro fertilization followed by embryo transfer), GIFT – gamete intrafallopian transfer, ICSI – intracytoplasmatic sperm injection − Draw and label simplified developmental schemes specified in a separate document. − Give examples of epigenetic mechanisms (at least three of them) and explain how these may affect the formation of phenotype. − Give examples of ethical issues in embryology (at least three of them). − Explain how the sperm cells are formed, starting with primordial gonocytes. Compare the nuclear DNA content, numbers of chromosomes, cell shape and size in all stages. − Explain how the Sertoli cells and Leydig cells contribute to spermatogenesis. − List the parameters used for sperm analysis. What are their normal values? − Explain how the mature oocytes differentiate, starting with oogonia. − Explain how the LH and FSH contribute to oogenesis. -

No Live Individual Homozygous for a Novel Endoglin Mutation Was Found in a Consanguineous Arab Family with Hereditary Haemorrhag
1of4 J Med Genet: first published as 10.1136/jmg.2004.022079 on 1 November 2004. Downloaded from ONLINE MUTATION REPORT No live individual homozygous for a novel endoglin mutation was found in a consanguineous Arab family with hereditary haemorrhagic telangiectasia A Karabegovic*, M Shinawi*, U Cymerman, M Letarte ............................................................................................................................... J Med Genet 2004;41:e119 (http://www.jmedgenet.com/cgi/content/full/41/11/e119). doi: 10.1136/jmg.2004.022079 ereditary haemorrhagic telangiectasia (HHT or Rendu- Osler-Weber syndrome; MIM 187300) is characterised Key points Hby vascular dysplasia and is inherited in an autosomal dominant manner. HHT occurs among many ethnic groups N Mutation analysis was performed in a large Arab over a wide geographical area. Recent epidemiological studies family with a known history of hereditary haemor- have revealed an incidence for this disease of 1 in 5000– rhagic telangiectasia (HHT) and consanguinity. 12 8000. In most cases, the manifestations of HHT are not N A novel exon 7 missense mutation (c.932TRG) in the present at birth, but develop with age; epistaxis is usually the Endoglin (ENG) gene was found in the proband, earliest sign, often occurring in childhood, while mucocuta- suggesting HHT1. neous and gastrointestinal telangiectases develop progres- sively with age.3 Arteriovenous malformations (AVMs) in the N The mutation was present as a single allele in ten pulmonary, cerebral, or hepatic circulations account for some relatives with clinical signs of disease but was absent of the most devastating clinical complications of HHT and are from 21 unaffected family members, indicating that the due to direct connections between arteries and veins.4 The mutation segregates with the phenotype. -

In Situ Detection of Tbx5 Expression in Developing Chick Embryonic Heart
San Jose State University SJSU ScholarWorks Master's Theses Master's Theses and Graduate Research Fall 2010 In Situ Detection Of Tbx5 Expression In Developing Chick Embryonic Heart Vaishali Agarwal San Jose State University Follow this and additional works at: https://scholarworks.sjsu.edu/etd_theses Recommended Citation Agarwal, Vaishali, "In Situ Detection Of Tbx5 Expression In Developing Chick Embryonic Heart" (2010). Master's Theses. 3901. DOI: https://doi.org/10.31979/etd.84mn-c4vy https://scholarworks.sjsu.edu/etd_theses/3901 This Thesis is brought to you for free and open access by the Master's Theses and Graduate Research at SJSU ScholarWorks. It has been accepted for inclusion in Master's Theses by an authorized administrator of SJSU ScholarWorks. For more information, please contact [email protected]. IN SITU DETECTION OF Tbx 5 EXPRESSION IN DEVELOPING EMBRYONIC CHICK HEART A Thesis Presented to The Faculty of the Department of Biological Sciences San Jose State University In Partial Fulfillment of the Requirements for the Degree Master of Science by Vaishali Agarwal December 2010 © 2010 Vaishali Agarwal ALL RIGHTS RESERVED The Designated Thesis Committee Approves the Thesis Titled IN SITU DETECTION OF Tbx 5 EXPRESSION IN DEVELOPING EMBRYONIC CHICK HEART by Vaishali Agarwal APPROVED FOR THE DEPARTMENT OF BIOLOGICAL SCIENCES SAN JOSE STATE UNIVERSITY December 2010 Dr. Steven White Department of Biological Sciences Dr. Michael Sneary Department of Biological Sciences Dr. Bob Fowler Department of Biological Sciences ABSTRACT IN SITU DETECTION OF Tbx 5 EXPRESSION IN DEVELOPING EMBRYONIC CHICK HEART by Vaishali Agarwal The Tbx 5 gene codes for a highly conserved transcription factor containing a DNA-binding motif called the T box (or T-domain). -

(Microsoft Powerpoint
6.1. DEVELOPMENT OF MESODERMAL ORGANS Pavel Krejci In vertebrates, the mesoderm becomes partitioned at an early stage into four zones, from medial to lateral: 1. NOTOCHORD : occupies the midline. 2. PARAXIAL MESODERM : future somites. 3. INTERMEDIATE MESODERM : forms gonads, kidneys, and adrenals. 4. LATERAL PLATE MESODERM : the lateral plate is subdivided by the coelom into the outer SOMATIC MESODERM (future limb buds) and inner SPLANCHNIC MESODERM that forms mesenteries and heart. The skeleton originates from three regions: Skull is formed from neural crest ; the vertebrae are formed from somites ; and the bones of the limbs are formed from limb buds and associated lateral plate . SOMITOGENESIS AND MYOGENESIS Somite patterning is a clearest example of segmental arrangement of the vertebrate body. Somites arise in anteroposterior sequence from the paraxial mesoderm by the action of forkhead transcription factors FoxC1 and C2. Somites start as loose cell associations called somitomeres, that later condense into the epithelial somites . This structure is transient as it undergoes epithelial-to- mesenchymal transformation to form the sclerotome (future vertebrae/ribs). Dorsal part of sclerotome forms tendons whereas the lateral part forms dermamyotome that later forms skin (dermatome) and muscles (myotome). Epaxial myotome forms segmental muscles of the body axis, hypaxial myotome forms muscles of the ventral body wall, limbs, and diaphragm. SEGMENTATION MECHANISM: Somites generated by molecular oscillator (a clock) operating in conjunction with a spatial gradient. One cycle of the clock forms one somite, the gradient determines that the somites are formed in anterior to posterior sequence. The clock represents a periodic expression of c-hairy 1, that encodes for transcription factor bHLH in chick. -

Investigating a Microrna-499-5P Network During Cardiac Development
Investigating a microRNA-499-5p network during cardiac development Thesis for a PhD degree Submitted to University of East Anglia by Johannes Gottfried Wittig This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that use of any information derived therefrom must be in accordance with current UK Copyright Law. In addition, any quotation or extract must include full attribution. Principal Investigator: Prof. Andrea Münsterberg Submission Date: 10.05.2019 Declaration of own work Declaration of own work I, Johannes Wittig, confirm that the work for the report with the title: “Investigating a microRNA-499-5p network during cardiac development” was undertaken by myself and that no help was provided from other sources than those allowed. All sections of the report that use quotes or describe an argument or development investigated by other scientist have been referenced, including all secondary literature used, to show that this material has been adopted to support my report. Place/Date Signature II Acknowledgements Acknowledgements I am very happy that I had the chance to be part of the Münsterberg-lab for my PhD research, therefore I would very much like to thank Andrea Münsterberg for offering me this great position in her lab. I especially want to thank her for her patience with me in all the moments where I was impatient and complained about slow progress. I also would like to say thank you for the incredible freedom I had during my PhD work and the support she gave me in the lab but also the understanding for all my non-science related activities. -

Echocardiographic Evaluation of Anatomical Abnormalities Of
Jemds.com Original Research Article Echocardiographic Evaluation of Anatomical Abnormalities of Interventricular Septum (with Special Reference to Ventricular Septal Defect) in the Population of West Bengal between 1 and 12 Years of Age Arpan Kumar Goswami1, Dhruba Mandal2, Shruti Goswami3, Biswajit Majumdar4, Adrija Mandal5 1Assistant Professor, Department of Anatomy, Bankura Sammilani Medical College, Bankura, West Bengal, India. 2Associate Professor, Department of Anatomy, Bankura Sammilani Medical College, Bankura, West Bengal, India. 3Scholar, Tripura Medical College, Agartala, Tripura, India. 4Associate Professor Department of Cardiology, R.G. Kar Medical College, Kolkata, West Bengal, India. 5Scholar, Burdwan Model School, Purba Bardhaman, West Bengal, India. ABSTRACT BACKGROUND Right and left ventricles of human heart are separated by interventricular septum. Corresponding Author: The septum develops in early embryonic period from three sources: primitive Dr. Dhruba Mandal, Rabindrapalli, ventricular septum, bulbar septum and endocardial cushion. Primitive ventricular Burdwan-713101, septum forms the major part of muscular septum, some contribution comes from West Bengal, India. bulbar septum. Endocardial cushion forms the membranous part of ventricular E-mail: [email protected] septum. Obviously, these three parts fuse to form a complete septum. Sometimes they do not fuse properly, or one of them does not proliferate properly, leading to DOI: 10.14260/jemds/2019/777 formation of a defective septum containing one or more foramina. This condition is called ventricular septal defect. Commonly, endocardial cushion does not proliferate Financial or Other Competing Interests: properly producing defect in membranous septum. Less commonly foramina are None. situated in muscular septum. Perimembranous VSD is described as foramen lies in How to Cite This Article: membranous septum and surrounding it, which is due to non-proliferation of Goswami AK, Mandal D, Goswami S, et al. -
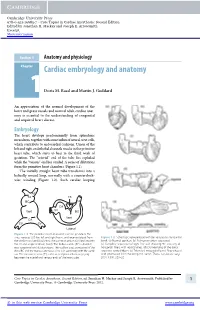
Core Topics in Cardiac Anesthesia, Second Edition, Ed
Cambridge University Press 978-0-521-19685-7 - Core Topics in Cardiac Anesthesia: Second Edition Edited by Jonathan H. Mackay and Joseph E. Arrowsmith Excerpt More information Section 1 Anatomy and physiology Chapter1 Cardiac embryology and anatomy Doris M. Rassl and Martin J. Goddard An appreciation of the normal development of the heart and great vessels and normal adult cardiac anat- omy is essential to the understanding of congenital and acquired heart disease. Embryology The heart develops predominantly from splanchnic mesoderm, together with some influx of neural crest cells, which contribute to endocardial cushions. Union of the left and right endothelial channels results in the primitive heart tube, which starts to beat in the third week of gestation. The “arterial” end of the tube lies cephalad while the “venous” end lies caudad. A series of dilatations form the primitive heart chambers (Figure 1.1). The initially straight heart tube transforms into a helically wound loop, normally with a counterclock- wise winding (Figure 1.2). Such cardiac looping AS TA TS BC TA CA Vent SV BC Vent CA SV SV Lateral Figure 1.1 The primitive heart at around 3 weeks’ gestation. The sinus venosus (SV) has left and right horns, and receives blood from Figure 1.2 Schematic representation of the ventricular myocardial the vitelline and umbilical veins. The common atrium (CA) lies between band. (a) Normal position. (b) Pulmonary artery separated. the SV and single ventricle (Vent). The bulbus cordis (BC) is divided (c) Complete separation of right free wall showing 90 crossing of into a proximal and distal portions. -

The Sinus Venosus Typeof Interatrial Septal Defect*
Thorax: first published as 10.1136/thx.13.1.12 on 1 March 1958. Downloaded from Thorax (I9%8), 13, 12. THE SINUS VENOSUS TYPE OF INTERATRIAL SEPTAL DEFECT* BY H. R. S. HARLEY Cardiff (RECEIVED FOR PUBLICATION DECEMBER 30, 1957) Defects of the interatrial septum, other than namely, (1) it lies above and independent of valvular patency of the foramen ovale, are often the fossa ovalis; (2) its margin is incomplete, classified into ostium primum and ostium secun- being absent superiorly and incomplete pos- dum varieties. The relationship of the former type teriorly; and (3) it is associated with anomalous to abnormal development of the atrioventricular drainage of the right superior, and sometimes of canal has been stressed by several workers, includ- the right middle or inferior, pulmonary vein. This ing Rogers and Edwards (1948), Wakai and type of defect is illustrated in Fig. 1 (after Lewis Edwards (1956), Wakai, Swan, and Wood (1956), et al., 1955) and Fig. 2 (after Geddes, 1912). In Brandenburg and DuShane (1956), Toscano- the case reported by Ross (1956), who kindly per- Barbosa, Brandenburg, and Burchell (1956), and mitted me to see the heart, the interatrial Cooley and Kirklin (1956). These workers prefer communication was described as ". lying the term "persistent common within the orifice of atrioventricular the superior vena cava in itscopyright. canal " to "persistent ostium primum." medial wall opposite the mouths of the anomalous In addition to the above types of interatrial pulmonary veins." Ross goes on to say: "On septal defect there is a third variety, which was casual inspection of the interior of the left atrium, described as long ago as 1868 by Wagstaffe, but the defect was not visible unless a search was made which has come into prominence only since the within the superior caval orifice." The relation- http://thorax.bmj.com/ introduction of surgical repair of interatrial ship of the defect to the orifice of the superior communications under direct vision. -

Cardiac Development
2 Cardiac Development BRAD J. MARTINSEN, PhD AND JAMIE L. LOHR, MD CONTENTS INTRODUCTION TO HUMAN HEART EMBRYOLOGY AND DEVELOPMENT PRIMARY HEART FIELD AND LINEAR HEART TUBE FORMATION SECONDARY HEART FIELD, OUTFLOWTRACT FORMATION, AND CARDIAC LOOPING CARDIAC NEURAL CREST AND OUTFLOWTRACT AND ATRIAL AND VENTRICULARSEPTATION PROEPICARDIUM AND CORONARY ARTERY DEVELOPMENT CARDIAC MATURATION SUMMARY OF EMBRYONIC CONTRIBUTIONTO HEART DEVELOPMENT REFERENCES 1. INTRODUCTION TO HUMAN HEART human congenital heart disease (5). In addition, great strides EMBRYOLOGY AND DEVELOPMENT have also been made in our knowledge of the contribution of the cardiac neural crest and the epicardium to overall heart devel- The primary heart field, secondary heart field, cardiac neural opment. crest, and proepicardium are the four major embryonic regions involved in the process of vertebrate heart develop- 2. PRIMARY HEART FIELD ment (Fig. 1). They each make an important contribution to AND LINEAR HEART TUBE FORMATION overall cardiac development, which occurs with complex developmental timing and regulation. This chapter describes The cells that will become the heart are among the first cell how these regions interact to form the final structure of the heart lineages formed in the vertebrate embryo (6,7). By day 15 of in relationship to the generalized developmental timeline of human development, the primitive streak has formed (8), and human embryology (Table 1). the first mesodermal cells to migrate (gastrulate) through the The heart is the first organ to fully form and function during primitive streak are cells fated to become the heart (9,10) vertebrate development, and many of the underlying mecha- (Fig. 2). These mesodermal cells migrate to an anterior and nisms are considered molecularly and developmentally con- lateral position where they form bilateral primary heart fields served (1). -

6. Heart and Circulatory System I
6. HEART AND CIRCULATORY SYSTEM I Dr. Taube P. Rothman P&S 12-520 [email protected] 212-305-7930 RECOMMENDED READING: Larsen Human Embryology, 3rd Edition, pp. 195-199; 157-169 top left; 172-174; bottom 181-182; 187-top 189, Simbryo-cardiovascular system SUMMARY: The circulatory system, consisting of heart, blood vessels, and blood cells is the first functional organ to develop. This lecture will focus on the formation of the embryonic vasculature, the origin and formation of the early heart tube and primitive cardiac chambers, cardiac looping, and the primitive circulation. Between the 5th - 8th week of embryonic development, the tubular heart is remodeled into a four chambered structure. We will see how right and left atrioventricular canals connect each atrium with its respective ventricle, and how the atrial septum and definitive right and left atria form. We will also see why the great veins deliver blood to the right atrium while the pulmonary veins empty into the left. GLOSSARY: Angioblasts: precursors of blood vessels Angiogenesis: lengthening, branching, sprouting and remodeling of embryonic blood vessels Aortic arches: paired arteries surrounding the pharynx; portions will contribute to formation of the great arterial vessels Blood islands: clusters of cells in the yolk sac, connecting stalk and chorionic villi that form primitive blood vessels Cardiac jelly: gelatinous extracellular matrix that forms the middle layer of the heart tube Ductus venosus: shunts most of the blood in the umbilical vein into the inferior vena cava -

Cardiovascular Development
Cardiovascular development Dawei Dong PHD Meg Sleeper VMD Lecture goals • Understand developmental anomalies which result in congenital heart disease. • Correlation with clinical veterinary medicine. – Common congenital heart disease in veterinary patients. Know the developmental structures that are associated with congenital heart disease. 1 Lectrue 1: Development of the Blood Cells and the Heart LEARNING GOALS 1. explain the early development of the cardiac tube from visceral mesoderm ahead of the neural plate which is then folded beneath the pharynx of the head fold. 2. outline the fusion of the cardiac tube to form the simple linear heart, the segmentation and the loop formation with sinus venosus, atrium, ventricle, bulbus cordis, truncus arteriosus. 3. define the three circulatory arcs of the heart to/from the body tissues, the yolk sac (vitelline) and the allantois (umbilical) and describe their functions 4. describe the three periods of blood cell formation related to the yolk sac, the liver and the bone marrow. 5. understand the developmental process by which the conus cordis and truncus arteriosus are adapted to give the aortic and pulmonary trunk, i.e., the heart outlet separation. 6. show how septum formation in the primitive linear heart allows separate pumping of blood into the aortic and the pulmonary trunk. 7. describe the heart inlet separation through the incorporation of the sinus venosus and the pulmonary veins into the atrium. Early development of the heart The cardiac tube folds under the gut tube…… The cardiac tube(6) is established in the early gastrula as regions of visceral mesoderm ahead of the embryo itself.