Investigating a Microrna-499-5P Network During Cardiac Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Embryology and Anatomy of Fetal Heart
Prof. Saeed Abuel Makarem Dr. Jamila El Medany Objectives • By the end of this lecture the student should be able to: • Describe the formation, sit, union divisions of the of the heart tubes. • Describe the formation and fate of the sinus venosus. • Describe the partitioning of the common atrium and common ventricle. • Describe the partitioning of the truncus arteriosus. • List the most common cardiac anomalies. • The CVS is the first major system to function in the embryo. • The heart begins to beat at (22nd – 23rd ) days. • Blood flow begins during the beginning of the fourth week and can be visualized by Ultrasound Doppler Notochord: stimulates neural tube formation Somatic mesoderm Splanchnic mesoderm FORMATION OF THE HEART TUBE • The heart is the first functional organ to develop. • It develops from Splanchnic Mesoderm in the wall of the yolk sac (Cardiogenic Area): Cranial to the developing Mouth & Nervous system and Ventral to the developing Pericardial sac. • The heart primordium is first evident at day 18 (as an Angioplastic cords which soon canalize to form the 2 heart tubes). • As the Head Fold completed, the developing heart tubes change their position and become in the Ventral aspect of the embryo, Dorsal to the developing Pericardial sac. • . Development of the Heart tube • After Lateral Folding of the embryo, the 2 heart tubes approach each other and fuse to form a single Endocardial Heart tube within the pericardial sac. • Fusion of the two tubes occurs in a Craniocaudal direction. What is the • The heart tube grows faster than shape of the the pericardial sac, so it shows 5 alternate dilations separated by Heart Tube? constrictions. -

Goals and Outcomes – Gametogenesis, Fertilization (Embryology Chapter 1)
Department of Histology and Embryology, Faculty of Medicine in Pilsen, Charles University, Czech Republic; License Creative Commons - http://creativecommons.org/licenses/by-nc-nd/3.0/ Goals and outcomes – Gametogenesis, fertilization (Embryology chapter 1) Be able to: − Define and use: progenesis, gametogenesis, primordial gonocytes, spermatogonia, primary and secondary spermatocytes, spermatids, sperm cells (spermatozoa), oogonia, primary and secondary oocytes, polar bodies, ovarian follicles (primordial, primary, secondary, tertiary), membrane granulosa, cumulus oophorus, follicular antrum, theca folliculi interna and externa, zona pellucida, corona radiata, ovulation, corpus luteum, corpus albicans, follicular atresia, expanded cumulus, luteinizing hormone (LH), follicle-stimulating hormone (FSH), human chorionic gonadotropin (hCG), sperm capacitation, acrosome reaction, cortical reaction and zona reaction, fertilization, zygote, cleavage, implantation, gastrulation, organogenesis, embryo, fetus, cell division, differentiation, morphogenesis, condensation, migration, delamination, apoptosis, induction, genotype, phenotype, epigenetics, ART – assisted reproductive techniques, spermiogram, IVF-ET (in vitro fertilization followed by embryo transfer), GIFT – gamete intrafallopian transfer, ICSI – intracytoplasmatic sperm injection − Draw and label simplified developmental schemes specified in a separate document. − Give examples of epigenetic mechanisms (at least three of them) and explain how these may affect the formation of phenotype. − Give examples of ethical issues in embryology (at least three of them). − Explain how the sperm cells are formed, starting with primordial gonocytes. Compare the nuclear DNA content, numbers of chromosomes, cell shape and size in all stages. − Explain how the Sertoli cells and Leydig cells contribute to spermatogenesis. − List the parameters used for sperm analysis. What are their normal values? − Explain how the mature oocytes differentiate, starting with oogonia. − Explain how the LH and FSH contribute to oogenesis. -

Supplementary Information
Supplementary Information This text file includes: Supplementary Methods Supplementary Figure 1-13, 15-30 Supplementary Table 1-8, 16, 20-21, 23, 25-37, 40-41 1 1. Samples, DNA extraction and genome sequencing 1.1 Ethical statements and sample storage The ethical statements of collecting and processing tissue samples for each species are listed as follows: Myotis myotis: All procedures were carried out in accordance with the ethical guidelines and permits (AREC-13-38-Teeling) delivered by the University College Dublin and the Préfet du Morbihan, awarded to Emma Teeling and Sébastien Puechmaille respectively. A single M. myotis individual was humanely sacrificed given that she had lethal injuries, and dissected. Rhinolophus ferrumequinum: All the procedures were conducted under the license (Natural England 2016-25216-SCI-SCI) issued to Gareth Jones. The individual bat died unexpectedly and suddenly during sampling and was dissected immediately. Pipistrellus kuhlii: The sampling procedure was carried out following all the applicable national guidelines for the care and use of animals. Sampling was done in accordance with all the relevant wildlife legislation and approved by the Ministry of Environment (Ministero della Tutela del Territorio e del Mare, Aut.Prot. N˚: 13040, 26/03/2014). Molossus molossus: All sampling methods were approved by the Ministerio de Ambiente de Panamá (SE/A-29-18) and by the Institutional Animal Care and Use Committee of the Smithsonian Tropical Research Institute (2017-0815-2020). Phyllostomus discolor: P. discolor bats originated from a breeding colony in the Department Biology II of the Ludwig-Maximilians-University in Munich. Approval to keep and breed the bats was issued by the Munich district veterinary office. -

Applying Machine Learning to Breast Cancer Gene Expression Data to Predict Survival Likelihood Pegah Tavangar Thesis Submitted T
Applying Machine Learning to Breast Cancer Gene Expression Data to Predict Survival Likelihood Pegah Tavangar Thesis submitted to the University of Ottawa in partial Fulfillment of the requirements for the Master of Science in Chemistry and Biomolecular Sciences Department of Chemistry and Biomolecular Sciences Faculty of Science University of Ottawa © Pegah Tavangar, Ottawa, Canada, 2020 Abstract Analyzing the expression level of thousands of genes will provide additional information beneficial in improving cancer therapy or synthesizing a new drug. In this project, the expression of 48807 genes from primary human breast tumors cells was analyzed. Humans cannot make sense of such a large volume of gene expression data from each person. Therefore, we used Machine Learning as an automated system that can learn from the data and be able to predict results from the data. This project presents the use of Machine Learning to predict the likelihood of survival in breast cancer patients using gene expression profiling. Machine Learning techniques, such as Logistic Regression, Support Vector Machines, Random Forest, and different Feature Selection techniques were used to find essential genes that lead to breast cancer or help a patient to live longer. This project describes the evaluation of different Machine Learning algorithms to classify breast cancer tumors into two groups of high and low survival. ii Acknowledgments I would like to thank Dr. Jonathan Lee for providing me the opportunity to work with him on an exciting project. I would like to recognize the invaluable counsel that you all provided during my research. It was my honor to work with some other professors in the Faculty of Medicine, such as Dr. -

Product Datasheet INAVA Overexpression
Product Datasheet INAVA Overexpression Lysate NBP2-06832 Unit Size: 0.1 mg Store at -80C. Avoid freeze-thaw cycles. Protocols, Publications, Related Products, Reviews, Research Tools and Images at: www.novusbio.com/NBP2-06832 Updated 3/17/2020 v.20.1 Earn rewards for product reviews and publications. Submit a publication at www.novusbio.com/publications Submit a review at www.novusbio.com/reviews/destination/NBP2-06832 Page 1 of 2 v.20.1 Updated 3/17/2020 NBP2-06832 INAVA Overexpression Lysate Product Information Unit Size 0.1 mg Concentration The exact concentration of the protein of interest cannot be determined for overexpression lysates. Please contact technical support for more information. Storage Store at -80C. Avoid freeze-thaw cycles. Buffer RIPA buffer Target Molecular Weight 72.7 kDa Product Description Description Transient overexpression lysate of chromosome 1 open reading frame 106 (C1orf106), transcript variant 1 The lysate was created in HEK293T cells, using Plasmid ID RC215863 and based on accession number NM_018265. The protein contains a C-MYC/DDK Tag. Gene ID 55765 Gene Symbol C1ORF106 Species Human Notes HEK293T cells in 10-cm dishes were transiently transfected with a non-lipid polymer transfection reagent specially designed and manufactured for large volume DNA transfection. Transfected cells were cultured for 48hrs before collection. The cells were lysed in modified RIPA buffer (25mM Tris-HCl pH7.6, 150mM NaCl, 1% NP-40, 1mM EDTA, 1xProteinase inhibitor cocktail mix, 1mM PMSF and 1mM Na3VO4, and then centrifuged to clarify the lysate. Protein concentration was measured by BCA protein assay kit.This product is manufactured by and sold under license from OriGene Technologies and its use is limited solely for research purposes. -

No Live Individual Homozygous for a Novel Endoglin Mutation Was Found in a Consanguineous Arab Family with Hereditary Haemorrhag
1of4 J Med Genet: first published as 10.1136/jmg.2004.022079 on 1 November 2004. Downloaded from ONLINE MUTATION REPORT No live individual homozygous for a novel endoglin mutation was found in a consanguineous Arab family with hereditary haemorrhagic telangiectasia A Karabegovic*, M Shinawi*, U Cymerman, M Letarte ............................................................................................................................... J Med Genet 2004;41:e119 (http://www.jmedgenet.com/cgi/content/full/41/11/e119). doi: 10.1136/jmg.2004.022079 ereditary haemorrhagic telangiectasia (HHT or Rendu- Osler-Weber syndrome; MIM 187300) is characterised Key points Hby vascular dysplasia and is inherited in an autosomal dominant manner. HHT occurs among many ethnic groups N Mutation analysis was performed in a large Arab over a wide geographical area. Recent epidemiological studies family with a known history of hereditary haemor- have revealed an incidence for this disease of 1 in 5000– rhagic telangiectasia (HHT) and consanguinity. 12 8000. In most cases, the manifestations of HHT are not N A novel exon 7 missense mutation (c.932TRG) in the present at birth, but develop with age; epistaxis is usually the Endoglin (ENG) gene was found in the proband, earliest sign, often occurring in childhood, while mucocuta- suggesting HHT1. neous and gastrointestinal telangiectases develop progres- sively with age.3 Arteriovenous malformations (AVMs) in the N The mutation was present as a single allele in ten pulmonary, cerebral, or hepatic circulations account for some relatives with clinical signs of disease but was absent of the most devastating clinical complications of HHT and are from 21 unaffected family members, indicating that the due to direct connections between arteries and veins.4 The mutation segregates with the phenotype. -

In Situ Detection of Tbx5 Expression in Developing Chick Embryonic Heart
San Jose State University SJSU ScholarWorks Master's Theses Master's Theses and Graduate Research Fall 2010 In Situ Detection Of Tbx5 Expression In Developing Chick Embryonic Heart Vaishali Agarwal San Jose State University Follow this and additional works at: https://scholarworks.sjsu.edu/etd_theses Recommended Citation Agarwal, Vaishali, "In Situ Detection Of Tbx5 Expression In Developing Chick Embryonic Heart" (2010). Master's Theses. 3901. DOI: https://doi.org/10.31979/etd.84mn-c4vy https://scholarworks.sjsu.edu/etd_theses/3901 This Thesis is brought to you for free and open access by the Master's Theses and Graduate Research at SJSU ScholarWorks. It has been accepted for inclusion in Master's Theses by an authorized administrator of SJSU ScholarWorks. For more information, please contact [email protected]. IN SITU DETECTION OF Tbx 5 EXPRESSION IN DEVELOPING EMBRYONIC CHICK HEART A Thesis Presented to The Faculty of the Department of Biological Sciences San Jose State University In Partial Fulfillment of the Requirements for the Degree Master of Science by Vaishali Agarwal December 2010 © 2010 Vaishali Agarwal ALL RIGHTS RESERVED The Designated Thesis Committee Approves the Thesis Titled IN SITU DETECTION OF Tbx 5 EXPRESSION IN DEVELOPING EMBRYONIC CHICK HEART by Vaishali Agarwal APPROVED FOR THE DEPARTMENT OF BIOLOGICAL SCIENCES SAN JOSE STATE UNIVERSITY December 2010 Dr. Steven White Department of Biological Sciences Dr. Michael Sneary Department of Biological Sciences Dr. Bob Fowler Department of Biological Sciences ABSTRACT IN SITU DETECTION OF Tbx 5 EXPRESSION IN DEVELOPING EMBRYONIC CHICK HEART by Vaishali Agarwal The Tbx 5 gene codes for a highly conserved transcription factor containing a DNA-binding motif called the T box (or T-domain). -

(Microsoft Powerpoint
6.1. DEVELOPMENT OF MESODERMAL ORGANS Pavel Krejci In vertebrates, the mesoderm becomes partitioned at an early stage into four zones, from medial to lateral: 1. NOTOCHORD : occupies the midline. 2. PARAXIAL MESODERM : future somites. 3. INTERMEDIATE MESODERM : forms gonads, kidneys, and adrenals. 4. LATERAL PLATE MESODERM : the lateral plate is subdivided by the coelom into the outer SOMATIC MESODERM (future limb buds) and inner SPLANCHNIC MESODERM that forms mesenteries and heart. The skeleton originates from three regions: Skull is formed from neural crest ; the vertebrae are formed from somites ; and the bones of the limbs are formed from limb buds and associated lateral plate . SOMITOGENESIS AND MYOGENESIS Somite patterning is a clearest example of segmental arrangement of the vertebrate body. Somites arise in anteroposterior sequence from the paraxial mesoderm by the action of forkhead transcription factors FoxC1 and C2. Somites start as loose cell associations called somitomeres, that later condense into the epithelial somites . This structure is transient as it undergoes epithelial-to- mesenchymal transformation to form the sclerotome (future vertebrae/ribs). Dorsal part of sclerotome forms tendons whereas the lateral part forms dermamyotome that later forms skin (dermatome) and muscles (myotome). Epaxial myotome forms segmental muscles of the body axis, hypaxial myotome forms muscles of the ventral body wall, limbs, and diaphragm. SEGMENTATION MECHANISM: Somites generated by molecular oscillator (a clock) operating in conjunction with a spatial gradient. One cycle of the clock forms one somite, the gradient determines that the somites are formed in anterior to posterior sequence. The clock represents a periodic expression of c-hairy 1, that encodes for transcription factor bHLH in chick. -

Mouse Inava Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Inava Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Inava conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Inava gene (NCBI Reference Sequence: NM_028872.3 ; Ensembl: ENSMUSG00000041605 ) is located on Mouse chromosome 1. 10 exons are identified, with the ATG start codon in exon 1 and the TAA stop codon in exon 10 (Transcript: ENSMUST00000120339). Exon 2 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Inava gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-100N3 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 2 starts from about 10.04% of the coding region. The knockout of Exon 2 will result in frameshift of the gene. The size of intron 1 for 5'-loxP site insertion: 6240 bp, and the size of intron 2 for 3'-loxP site insertion: 609 bp. The size of effective cKO region: ~649 bp. The cKO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele gRNA region 5' gRNA region 3' 1 2 3 4 5 10 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Inava Homology arm cKO region loxP site Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Echocardiographic Evaluation of Anatomical Abnormalities Of
Jemds.com Original Research Article Echocardiographic Evaluation of Anatomical Abnormalities of Interventricular Septum (with Special Reference to Ventricular Septal Defect) in the Population of West Bengal between 1 and 12 Years of Age Arpan Kumar Goswami1, Dhruba Mandal2, Shruti Goswami3, Biswajit Majumdar4, Adrija Mandal5 1Assistant Professor, Department of Anatomy, Bankura Sammilani Medical College, Bankura, West Bengal, India. 2Associate Professor, Department of Anatomy, Bankura Sammilani Medical College, Bankura, West Bengal, India. 3Scholar, Tripura Medical College, Agartala, Tripura, India. 4Associate Professor Department of Cardiology, R.G. Kar Medical College, Kolkata, West Bengal, India. 5Scholar, Burdwan Model School, Purba Bardhaman, West Bengal, India. ABSTRACT BACKGROUND Right and left ventricles of human heart are separated by interventricular septum. Corresponding Author: The septum develops in early embryonic period from three sources: primitive Dr. Dhruba Mandal, Rabindrapalli, ventricular septum, bulbar septum and endocardial cushion. Primitive ventricular Burdwan-713101, septum forms the major part of muscular septum, some contribution comes from West Bengal, India. bulbar septum. Endocardial cushion forms the membranous part of ventricular E-mail: [email protected] septum. Obviously, these three parts fuse to form a complete septum. Sometimes they do not fuse properly, or one of them does not proliferate properly, leading to DOI: 10.14260/jemds/2019/777 formation of a defective septum containing one or more foramina. This condition is called ventricular septal defect. Commonly, endocardial cushion does not proliferate Financial or Other Competing Interests: properly producing defect in membranous septum. Less commonly foramina are None. situated in muscular septum. Perimembranous VSD is described as foramen lies in How to Cite This Article: membranous septum and surrounding it, which is due to non-proliferation of Goswami AK, Mandal D, Goswami S, et al. -
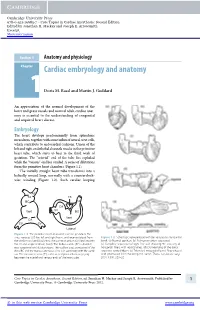
Core Topics in Cardiac Anesthesia, Second Edition, Ed
Cambridge University Press 978-0-521-19685-7 - Core Topics in Cardiac Anesthesia: Second Edition Edited by Jonathan H. Mackay and Joseph E. Arrowsmith Excerpt More information Section 1 Anatomy and physiology Chapter1 Cardiac embryology and anatomy Doris M. Rassl and Martin J. Goddard An appreciation of the normal development of the heart and great vessels and normal adult cardiac anat- omy is essential to the understanding of congenital and acquired heart disease. Embryology The heart develops predominantly from splanchnic mesoderm, together with some influx of neural crest cells, which contribute to endocardial cushions. Union of the left and right endothelial channels results in the primitive heart tube, which starts to beat in the third week of gestation. The “arterial” end of the tube lies cephalad while the “venous” end lies caudad. A series of dilatations form the primitive heart chambers (Figure 1.1). The initially straight heart tube transforms into a helically wound loop, normally with a counterclock- wise winding (Figure 1.2). Such cardiac looping AS TA TS BC TA CA Vent SV BC Vent CA SV SV Lateral Figure 1.1 The primitive heart at around 3 weeks’ gestation. The sinus venosus (SV) has left and right horns, and receives blood from Figure 1.2 Schematic representation of the ventricular myocardial the vitelline and umbilical veins. The common atrium (CA) lies between band. (a) Normal position. (b) Pulmonary artery separated. the SV and single ventricle (Vent). The bulbus cordis (BC) is divided (c) Complete separation of right free wall showing 90 crossing of into a proximal and distal portions. -

The Sinus Venosus Typeof Interatrial Septal Defect*
Thorax: first published as 10.1136/thx.13.1.12 on 1 March 1958. Downloaded from Thorax (I9%8), 13, 12. THE SINUS VENOSUS TYPE OF INTERATRIAL SEPTAL DEFECT* BY H. R. S. HARLEY Cardiff (RECEIVED FOR PUBLICATION DECEMBER 30, 1957) Defects of the interatrial septum, other than namely, (1) it lies above and independent of valvular patency of the foramen ovale, are often the fossa ovalis; (2) its margin is incomplete, classified into ostium primum and ostium secun- being absent superiorly and incomplete pos- dum varieties. The relationship of the former type teriorly; and (3) it is associated with anomalous to abnormal development of the atrioventricular drainage of the right superior, and sometimes of canal has been stressed by several workers, includ- the right middle or inferior, pulmonary vein. This ing Rogers and Edwards (1948), Wakai and type of defect is illustrated in Fig. 1 (after Lewis Edwards (1956), Wakai, Swan, and Wood (1956), et al., 1955) and Fig. 2 (after Geddes, 1912). In Brandenburg and DuShane (1956), Toscano- the case reported by Ross (1956), who kindly per- Barbosa, Brandenburg, and Burchell (1956), and mitted me to see the heart, the interatrial Cooley and Kirklin (1956). These workers prefer communication was described as ". lying the term "persistent common within the orifice of atrioventricular the superior vena cava in itscopyright. canal " to "persistent ostium primum." medial wall opposite the mouths of the anomalous In addition to the above types of interatrial pulmonary veins." Ross goes on to say: "On septal defect there is a third variety, which was casual inspection of the interior of the left atrium, described as long ago as 1868 by Wagstaffe, but the defect was not visible unless a search was made which has come into prominence only since the within the superior caval orifice." The relation- http://thorax.bmj.com/ introduction of surgical repair of interatrial ship of the defect to the orifice of the superior communications under direct vision.