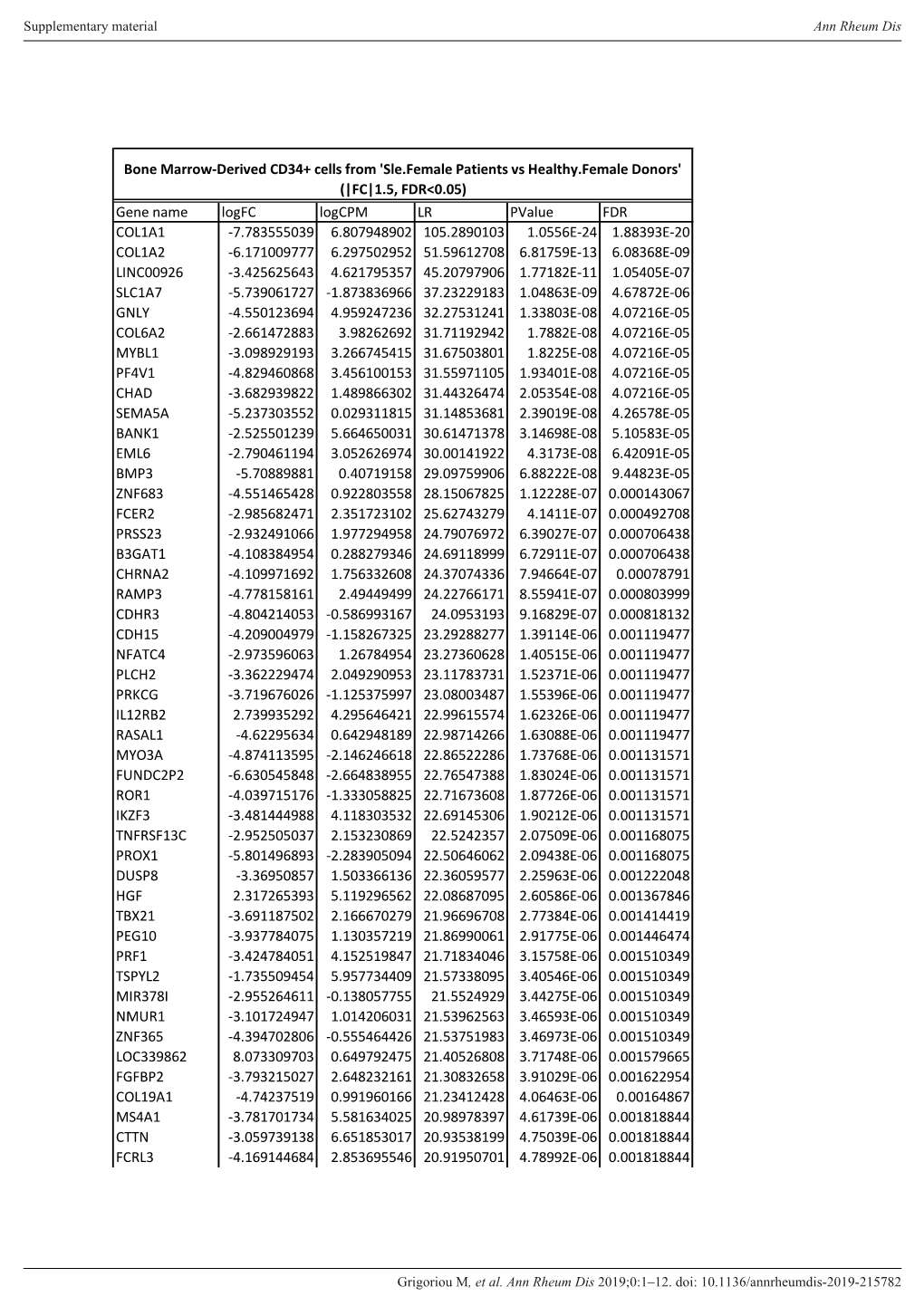
Gene Name Logfc Logcpm LR Pvalue FDR COL1A1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Database of Cattle Candidate Genes and Genetic Markers for Milk Production and Mastitis
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central doi:10.1111/j.1365-2052.2009.01921.x Database of cattle candidate genes and genetic markers for milk production and mastitis J. Ogorevc*, T. Kunej*, A. Razpet and P. Dovc Department of Animal Science, Biotechnical Faculty, University of Ljubljana, Domzale, Slovenia Summary A cattle database of candidate genes and genetic markers for milk production and mastitis has been developed to provide an integrated research tool incorporating different types of information supporting a genomic approach to study lactation, udder development and health. The database contains 943 genes and genetic markers involved in mammary gland development and function, representing candidates for further functional studies. The candidate loci were drawn on a genetic map to reveal positional overlaps. For identification of candidate loci, data from seven different research approaches were exploited: (i) gene knockouts or transgenes in mice that result in specific phenotypes associated with mam- mary gland (143 loci); (ii) cattle QTL for milk production (344) and mastitis related traits (71); (iii) loci with sequence variations that show specific allele-phenotype interactions associated with milk production (24) or mastitis (10) in cattle; (iv) genes with expression profiles associated with milk production (207) or mastitis (107) in cattle or mouse; (v) cattle milk protein genes that exist in different genetic variants (9); (vi) miRNAs expressed in bovine mammary gland (32) and (vii) epigenetically regulated cattle genes associated with mammary gland function (1). Fourty-four genes found by multiple independent analyses were suggested as the most promising candidates and were further in silico analysed for expression levels in lactating mammary gland, genetic variability and top biological func- tions in functional networks. -

G Protein Alpha 13 (GNA13) (NM 006572) Human Tagged ORF Clone Lentiviral Particle Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC207762L3V G protein alpha 13 (GNA13) (NM_006572) Human Tagged ORF Clone Lentiviral Particle Product data: Product Type: Lentiviral Particles Product Name: G protein alpha 13 (GNA13) (NM_006572) Human Tagged ORF Clone Lentiviral Particle Symbol: GNA13 Synonyms: G13 Vector: pLenti-C-Myc-DDK-P2A-Puro (PS100092) ACCN: NM_006572 ORF Size: 1131 bp ORF Nucleotide The ORF insert of this clone is exactly the same as(RC207762). Sequence: OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_006572.3 RefSeq Size: 4744 bp RefSeq ORF: 1134 bp Locus ID: 10672 UniProt ID: Q14344, A0A024R8M0 Domains: G-alpha Protein Families: Druggable Genome Protein Pathways: Long-term depression, Regulation of actin cytoskeleton, Vascular smooth muscle contraction MW: 44 kDa This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 G protein alpha 13 (GNA13) (NM_006572) Human Tagged ORF Clone Lentiviral Particle – RC207762L3V Gene Summary: Guanine nucleotide-binding proteins (G proteins) are involved as modulators or transducers in various transmembrane signaling systems (PubMed:15240885, PubMed:16787920, PubMed:16705036, PubMed:27084452). -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma
Anatomy and Pathology Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma Sarah L. Lake,1 Sarah E. Coupland,1 Azzam F. G. Taktak,2 and Bertil E. Damato3 PURPOSE. To detect deletions and loss of heterozygosity of disease is fatal in 92% of patients within 2 years of diagnosis. chromosome 3 in a rare subset of fatal, disomy 3 uveal mela- Clinical and histopathologic risk factors for UM metastasis noma (UM), undetectable by fluorescence in situ hybridization include large basal tumor diameter (LBD), ciliary body involve- (FISH). ment, epithelioid cytomorphology, extracellular matrix peri- ϩ ETHODS odic acid-Schiff-positive (PAS ) loops, and high mitotic M . Multiplex ligation-dependent probe amplification 3,4 5 (MLPA) with the P027 UM assay was performed on formalin- count. Prescher et al. showed that a nonrandom genetic fixed, paraffin-embedded (FFPE) whole tumor sections from 19 change, monosomy 3, correlates strongly with metastatic death, and the correlation has since been confirmed by several disomy 3 metastasizing UMs. Whole-genome microarray analy- 3,6–10 ses using a single-nucleotide polymorphism microarray (aSNP) groups. Consequently, fluorescence in situ hybridization were performed on frozen tissue samples from four fatal dis- (FISH) detection of chromosome 3 using a centromeric probe omy 3 metastasizing UMs and three disomy 3 tumors with Ͼ5 became routine practice for UM prognostication; however, 5% years’ metastasis-free survival. to 20% of disomy 3 UM patients unexpectedly develop metas- tases.11 Attempts have therefore been made to identify the RESULTS. Two metastasizing UMs that had been classified as minimal region(s) of deletion on chromosome 3.12–15 Despite disomy 3 by FISH analysis of a small tumor sample were found these studies, little progress has been made in defining the key on MLPA analysis to show monosomy 3. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Regulation of Transcription and Regulatory Networks for Muscle Growth * * * * A
Regulation Of Transcription And Regulatory Networks For Muscle Growth * * * * A. Reverter , N.J. Hudson , Q. Gu and B.P. Dalrymple Introduction The advent of microarray gene expression technology has provided animal scientists with an unprecedented ability to profile the transcriptional changes during skeletal muscle growth. With respect to meat quality, most of the effort has concentrated on the understanding of fat and energy metabolism (reviewed by Hausman et al . (2009)). Graugnard et al . (2009) explored the network among 31 genes associated with aspects of adipogenesis and energy metabolism in bovine skeletal muscle and in response to two distinct diets. Also, Freyssenet (2007) reviewed the roles that energy-sensing molecules and mitochondria have in the regulation of gene expression in muscle. However, other mechanisms such as cell cycle, glycolysis, extra-cellular matrix, ribosomal proteins and the immune system play a significant role in development, and this role can work in a tissue-specific manner. Hudson et al . (2009a) reported various functional modules underpinning the transcriptional regulation of bovine skeletal muscle. The authors integrated a total of six gene co-expression networks, each developed using the PCIT algorithm (Reverter and Chan (2008)), and proposed a Module-to-Regulator heuristic by which those transcription factors (TF) with the highest average absolute correlation co-expression with the genes present in each module are deemed to be the relevant regulators. However, this Module-to-Regulator approach failed to capture some well-known regulators of muscle fibre type composition, and the use of more sophisticated methods such as the differential wiring approach of Hudson et al . -

PLXNB1 (Plexin
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review PLXNB1 (plexin B1) José Javier Gómez-Román, Montserrat Nicolas Martínez, Servando Lazuén Fernández, José Fernando Val-Bernal Department of Anatomical Pathology, Marques de Valdecilla University Hospital, Medical Faculty, University of Cantabria, Santander, Spain (JJGR, MN, SL, JFVB) Published in Atlas Database: March 2009 Online updated version: http://AtlasGeneticsOncology.org/Genes/PLXNB1ID43413ch3p21.html DOI: 10.4267/2042/44702 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2010 Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Pseudogene No. Other names: KIAA0407; MGC149167; OTTHUMP00000164806; PLEXIN-B1; PLXN5; SEP Protein HGNC (Hugo): PLXNB1 Location: 3p21.31 Description Local order: The Plexin B1 gene is located between 2135 Amino acids (AA). Plexins are receptors for axon FBXW12 and CCDC51 genes. molecular guidance molecules semaphorins. Plexin signalling is important in pathfinding and patterning of both neurons and developing blood vessels. Plexin-B1 is a surface cell receptor. When it binds to its ligand SEMA4D it activates several pathways by binding of cytoplasmic ligands, like RHOA activation and subsequent changes of the actin cytoskeleton, axon guidance, invasive growth and cell migration. It monomers and heterodimers with PLXNB2 after proteolytic processing. Binds RAC1 that has been activated by GTP binding. It binds PLXNA1 and by similarity ARHGEF11, Note ARHGEF12, ERBB2, MET, MST1R, RND1, NRP1 Size: 26,200 bases. and NRP2. Orientation: minus strand. This family features the C-terminal regions of various plexins. The cytoplasmic region, which has been called DNA/RNA a SEX domain in some members of this family is involved in downstream signalling pathways, by Description interaction with proteins such as Rac1, RhoD, Rnd1 and other plexins. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

TTDN1 (MPLKIP) (NM 138701) Human 3' UTR Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for SC204348 TTDN1 (MPLKIP) (NM_138701) Human 3' UTR Clone Product data: Product Type: 3' UTR Clones Product Name: TTDN1 (MPLKIP) (NM_138701) Human 3' UTR Clone Vector: pMirTarget (PS100062) Symbol: MPLKIP Synonyms: ABHS; C7orf11; ORF20; TTD4 ACCN: NM_138701 Insert Size: 2000 bp This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 TTDN1 (MPLKIP) (NM_138701) Human 3' UTR Clone – SC204348 Insert Sequence: >SC204348 3’UTR clone of NM_138701 The sequence shown below is from the reference sequence of NM_138701. The complete sequence of this clone may contain minor differences, such as SNPs. Blue=Stop Codon Red=Cloning site GGCAAGTTGGACGCCCGCAAGATCCGCGAGATTCTCATTAAGGCCAAGAAGGGCGGAAAGATCGCCGTG TAACAATTGGCAGAGCTCAGAATTCAAGCGATCGCC ACAGGCAAAAAAGGAAGATACTTTTGTTAACATTTCTGAAATTCAACTGGAAGCTTCATGTGTCAGGAA CATCTTGGACAAAACTTTAAGTTGTGTTGATATAAATTTACCCAAAGATGATGACTTTGATTGGATAAT TAGTAAGGTCTTTTTGTTATTTTTCATCGTATCAGGTATTGTTGATATTAGAGAAAAAAGTAGGATAAC TTGCAACATTTAGCTCTGGAAGTACCTACCACATTTTAGAGATTTACCGTTTCCATATATTTAACATTC CTGGTTACATAATGGACATTTGTCTTTTAATGTTTTTTCAATGTTTTAAAATAAAACATTTTGTCTTCT AGCTATTGTGGTTTTGTGGTATGATAAAGAAGTAGACTTACTACAGTAATGCTTTGTAGTCACTTAGAG TTCATAGGTAAATGTTTTGCAAATTATTTTTGAAAATGAAATAGGTAAACCATCCTTTGAGCTGTAGAC -

The Role of High Density Lipoprotein Compositional and Functional Heterogeneity in Metabolic Disease
The role of high density lipoprotein compositional and functional heterogeneity in metabolic disease By Scott M. Gordon B.S. State University of New York College at Brockport October, 2012 A Dissertation Presented to the Faculty of The University of Cincinnati College of Medicine in partial fulfillment of the requirements for the Degree of Doctor of Philosophy from the Pathobiology and Molecular Medicine graduate program W. Sean Davidson Ph.D. (Chair) David Askew Ph.D. Professor and Thesis Chair Professor Department of Pathology Department of Pathology University of Cincinnati University of Cincinnati Francis McCormack M.D. Gangani Silva Ph.D. Professor Assistant Professor Department of Pathology Department of Pathology University of Cincinnati University of Cincinnati Jason Lu Ph.D. Assistant Professor Division of Bioinformatics Cincinnati Children’s Hospital i Abstract High density lipoproteins (HDL) are complexes of phospholipid, cholesterol and protein that circulate in the blood. Epidemiological studies have demonstrated a strong inverse correlation between plasma levels of HDL associated cholesterol (HDL-C) and the incidence of cardiovascular disease (CVD). Clinically, HDL-C is often measured and used in combination with low density lipoprotein cholesterol (LDL-C) to assess overall cardiovascular health. HDL have been shown to possess a wide variety of functional attributes which likely contribute to this protection including anti-inflammatory and anti- oxidative properties and the ability to remove excess cholesterol from peripheral tissues and deliver it to the liver for excretion, a process known as reverse cholesterol transport. This functional diversity might be explained by the complexity of HDL composition. Recent studies have taken advantage of advances in mass spectrometry technologies to characterize the proteome of total HDL finding that over 50 different proteins can associate with these particles. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of