Table Continues on Reverse Leukemia Inhibitory Factor Receptor (LIF-R) P42702 STAM-Binding Protein (STAMBP) O95630
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Table 1
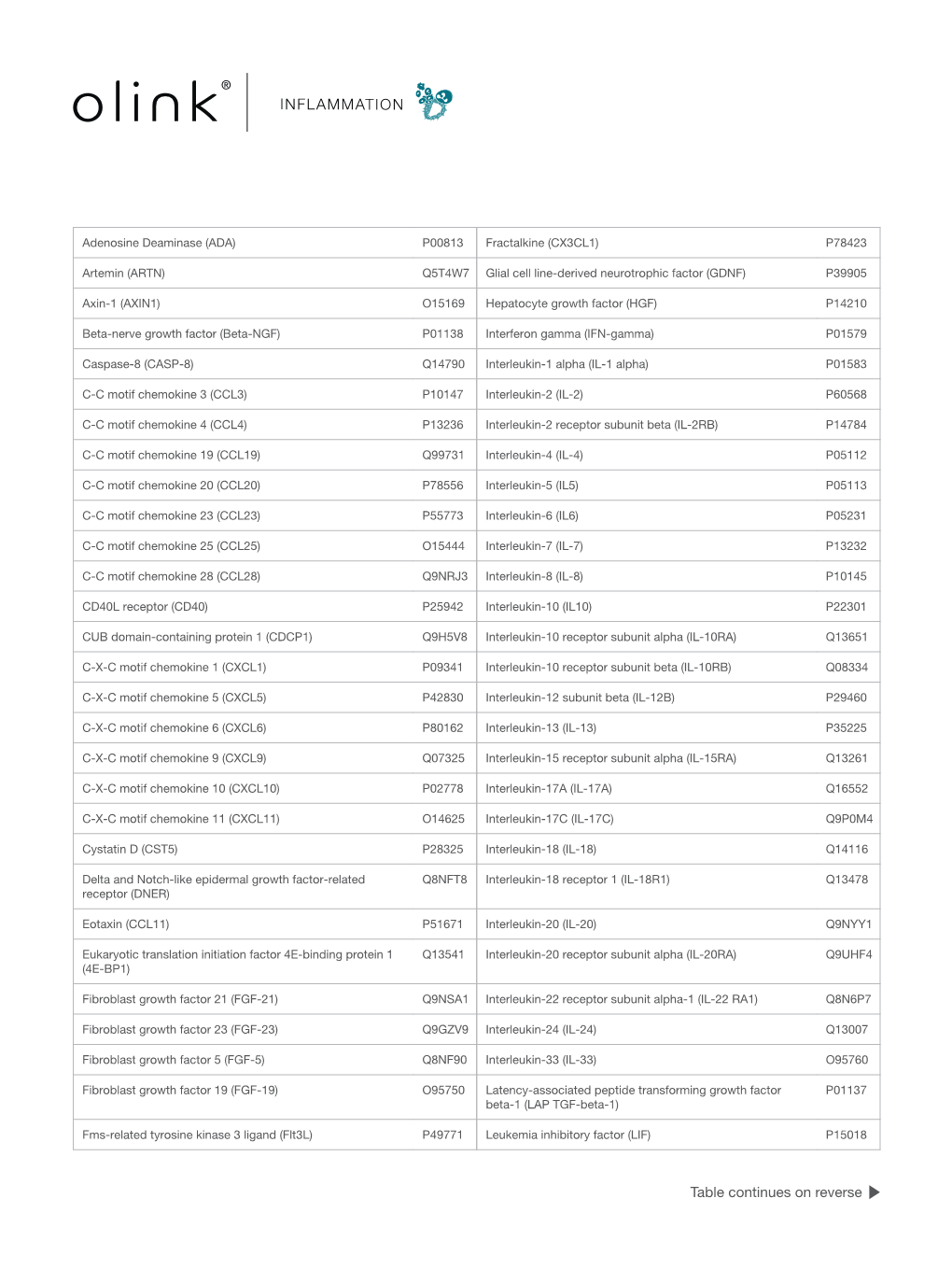
Supplementary table 1 List of the 92 proteins analyzed in the multiplex proximity extension assay (PEA) Long name (short name) UniProt No. LOD (pg/mL) Adenosine Deaminase (ADA) P00813 0.48 Artemin (ARTN) Q5T4W7 0.24 Axin-1 (AXIN1) O15169 61,0 Beta-nerve growth factor (Beta-NGF) P01138 0.48 Brain-derived neutrophic factor (BDNF) P23560 Caspase 8 (CASP-8) Q14790 0.48 C-C motif chemokine 4 (CCL4) P13236 1.9 C-C motif chemokine 19 (CCL19) Q99731 15,0 C-C motif chemokine 20 (CCL20) P78556 7.6 C-C motif chemokine 23 (CCL23) P55773 31,0 C-C motif chemokine 25 (CCL25) O15444 3.8 C-C motif chemokine 28 (CCL28) Q9NRJ3 61,0 CD40L receptor (CD40) P25942 0.01 CUB domain-containing protein 1 (CDCP1) Q9H5V8 0.12 C-X-C motif chemokine 1 (CXCL1) P09341 3.8 C-X-C motif chemokine 5 (CXCL5) P42830 0.95 C-X-C motif chemokine 6 (CXCL6) P80162 7.6 C-X-C motif chemokine 9 (CXCL9) Q07325 0.95 C-X-C motif chemokine 10 (CXCL10) P02778 7.6 C-X-C motif chemokine 11 (CXCL11) O14625 7.6 Cystatin D (CST5) P28325 1.9 Delta and Notch-like epidermal growth factor related receptor (DNER) Q8NFT8 0.95 Eotaxin-1 (CCL11) P51671 3.8 Eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1) Q13541 Fibroblast growth factor 5 (FGF-5) Q8NF90 1.9 Fibroblast growth factor 19 (FGF-19) O95750 7.6 Fibroblast growth factor 21 (FGF-21) Q9NSA1 31,0 Fibroblast growth factor 23 (FGF-23) Q9GZV9 122,0 Fms-related tyrosine kinase 3 ligand (FIt3L) P49771 0.01 Fractalkine (CX3CL1) P78423 15.3 Glial cell line-derived neutrophic factor (hGDNF) P39905 0.01 Hepatocyte growth factor (HGF) -

Artificial Liver Support Potential to Retard Regeneration?
REVIEW ARTICLE Artificial Liver Support Potential to Retard Regeneration? Emma J. Mullin, MBChB; Matthew S. Metcalfe, FRCS; Guy J. Maddern, MD Hypothesis: The concept of an “artificial liver” has been growth-promoting factors from these cultured hepato- in development for over 40 years. Such devices aim to cytes? temporarily assume metabolic and excretory functions of the liver, with removal of potentially hepatotoxic sub- Data Sources, Extraction, and Study Selection: stances, thereby clinically stabilizing patients and pre- Data were obtained using PubMed search for reports in- venting deterioration while awaiting transplantation. If volving liver support, extracorporeal circuits, dialysis, sufficient numbers of viable hepatocytes remain, regen- growth factors, and cytokines. Those reports specifi- eration and subsequent recovery of innate liver func- cally looking at the effect of artificial liver support on cy- tion may occur. However, these devices have not yet be- tokines and growth factors are discussed. come part of routine clinical use. Much less is known regarding the effect such devices have, if any, on circu- Conclusions: There is a paucity of information on the lating cytokines and growth factors and the subsequent key events and substances involved in hepatic regenera- effects on the regenerating liver. If these devices remove tion. In addition, there is a potential impact of liver sup- or reduce factors known to promote regeneration, is the port devices on the regeneration of substances associ- rate of regeneration retarded? Conversely, does the in- ated with hepatic regeneration. Further study is needed. corporation of hepatocytes into bioartificial support sys- tems confer an advantage through the production of Arch Surg. -

Human TGF Alpha ELISA Kit Basic Information: Catalog No.: UE1330 Size: 96T for Research Use Only
Efficient Professional Protein and Antibody Platforms Human TGF alpha ELISA Kit Basic information: Catalog No.: UE1330 Size: 96T For research use only. Not for diagnostic or therapeutic procedures. I. INTRODUCTION Transforming growth factor alpha (TGF-α) is upregulated in some human cancers. It is produced in macrophages, brain cells, and keratinocytes, and induces epithelial development. It is closely related to EGF, and can also bind to the EGF receptor with similar effects . TGFα stimulates neural cell proliferation in the adult injured brain. Transforming growth factor alpha gene (TGFA) maps to human chromosome 2 close to the breakpoint of the t (2;8) variant translocation in Burkitt lymphoma. Synthetic TGF-alpha was as active as murine epidermal growth factor in binding to the epidermal growth factor receptor and in stimulation of anchorage-dependent and of anchorage-independent growth of normal indicator cells in culture. Synthetic TGF-alpha stimulated plasminogen activator production in A 431 and HeLa cells; the stimulation was similar to that induced by epidermal growth factor. Furthermore, synthetic human TGF-alpha showed similar immunoreactivity when compared with rat TGF-alpha. Thus, the 50-amino acid TGF-alpha is likely to be the bioactive principle produced and secreted by tumor cell lines. II. ASSAY PRINCIPLES The Gene Universal Human TGF alpha ELISA (Enzyme-Linked Immunosorbent Assay) kit is an in vitro enzyme-linked immunosorbent assay for the quantitative measurement of Human TGF alpha in Cell Culture Supernatants, Serum, Plasma. This assay employs an antibody specific for Human TGF alpha coated on a 96-well plate. Standards and samples are pipetted into the wells and TGF alpha present in a sample is bound to the wells by the immobilized antibody. -

Molecular Mediators of Acute and Chronic Itch in Mouse and Human Sensory Neurons Manouela Valtcheva Washington University in St
Washington University in St. Louis Washington University Open Scholarship Arts & Sciences Electronic Theses and Dissertations Arts & Sciences Spring 5-15-2018 Molecular Mediators of Acute and Chronic Itch in Mouse and Human Sensory Neurons Manouela Valtcheva Washington University in St. Louis Follow this and additional works at: https://openscholarship.wustl.edu/art_sci_etds Part of the Neuroscience and Neurobiology Commons Recommended Citation Valtcheva, Manouela, "Molecular Mediators of Acute and Chronic Itch in Mouse and Human Sensory Neurons" (2018). Arts & Sciences Electronic Theses and Dissertations. 1596. https://openscholarship.wustl.edu/art_sci_etds/1596 This Dissertation is brought to you for free and open access by the Arts & Sciences at Washington University Open Scholarship. It has been accepted for inclusion in Arts & Sciences Electronic Theses and Dissertations by an authorized administrator of Washington University Open Scholarship. For more information, please contact [email protected]. WASHINGTON UNIVERSITY IN ST. LOUIS Division of Biology and Biomedical Sciences Neurosciences Dissertation Examination Committee: Robert W. Gereau, IV, Chair Yu-Qing Cao Sanjay Jain Qin Liu Durga Mohapatra Molecular Mediators of Acute and Chronic Itch in Mouse and Human Sensory Neurons by Manouela Vesselinova Valtcheva A dissertation presented to The Graduate School of Washington University in partial fulfillment of the requirements for the degree of Doctor of Philosophy May 2018 St. Louis, Missouri © 2018, Manouela V. Valtcheva Table -

Angiocrine Endothelium: from Physiology to Cancer Jennifer Pasquier1,2*, Pegah Ghiabi2, Lotf Chouchane3,4,5, Kais Razzouk1, Shahin Rafi3 and Arash Rafi1,2,3
Pasquier et al. J Transl Med (2020) 18:52 https://doi.org/10.1186/s12967-020-02244-9 Journal of Translational Medicine REVIEW Open Access Angiocrine endothelium: from physiology to cancer Jennifer Pasquier1,2*, Pegah Ghiabi2, Lotf Chouchane3,4,5, Kais Razzouk1, Shahin Rafi3 and Arash Rafi1,2,3 Abstract The concept of cancer as a cell-autonomous disease has been challenged by the wealth of knowledge gathered in the past decades on the importance of tumor microenvironment (TM) in cancer progression and metastasis. The sig- nifcance of endothelial cells (ECs) in this scenario was initially attributed to their role in vasculogenesis and angiogen- esis that is critical for tumor initiation and growth. Nevertheless, the identifcation of endothelial-derived angiocrine factors illustrated an alternative non-angiogenic function of ECs contributing to both physiological and pathological tissue development. Gene expression profling studies have demonstrated distinctive expression patterns in tumor- associated endothelial cells that imply a bilateral crosstalk between tumor and its endothelium. Recently, some of the molecular determinants of this reciprocal interaction have been identifed which are considered as potential targets for developing novel anti-angiocrine therapeutic strategies. Keywords: Angiocrine, Endothelium, Cancer, Cancer microenvironment, Angiogenesis Introduction of blood vessels in initiation of tumor growth and stated Metastatic disease accounts for about 90% of patient that in the absence of such angiogenesis, tumors can- mortality. Te difculty in controlling and eradicating not expand their mass or display a metastatic phenotype metastasis might be related to the heterotypic interaction [7]. Based on this theory, many investigators assumed of tumor and its microenvironment [1]. -

Artemin, a Novel Member of the GDNF Ligand Family, Supports Peripheral and Central Neurons and Signals Through the GFR␣3–RET Receptor Complex
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Neuron, Vol. 21, 1291±1302, December, 1998, Copyright 1998 by Cell Press Artemin, a Novel Member of the GDNF Ligand Family, Supports Peripheral and Central Neurons and Signals through the GFRa3±RET Receptor Complex Robert H. Baloh,* Malu G. Tansey,² et al., 1998; Milbrandt et al., 1998; reviewed by Grondin Patricia A. Lampe,² Timothy J. Fahrner,* and Gash, 1998). However, the GDNF ligands also influ- Hideki Enomoto,* Kelli S. Simburger,* ence a broad spectrum of other neuronal populations Melanie L. Leitner,² Toshiyuki Araki,* in both the CNS and PNS. GDNF and NTN both support Eugene M. Johnson, Jr.,² the survival of many peripheral neurons in culture, in- and Jeffrey Milbrandt*³ cluding sympathetic, parasympathetic, sensory, and en- *Department of Pathology and teric neurons (Buj-Bello et al., 1995; Ebendal et al., 1995; Department of Internal Medicine Trupp et al., 1995; Kotzbauer et al., 1996; Heuckeroth ² Department of Neurology and et al., 1998). In contrast, PSP does not share any of Department of Molecular Biology these peripheral activities but does support the survival and Pharmacology of dopaminergic midbrain neurons and motor neurons Washington University (Milbrandt et al., 1998). Despite the fact that GDNF acts School of Medicine on many cell populations in vitro, analysis of GDNF St. Louis, Missouri 63110 knockout mice revealed that the major developmental importance of GDNF is in the enteric nervous system and in kidney organogenesis, both of which are lost in Summary GDNF null mice (Moore et al., 1996; Pichel et al., 1996; Sanchez et al., 1996). -

Intravitreal Co-Administration of GDNF and CNTF Confers Synergistic and Long-Lasting Protection Against Injury-Induced Cell Death of Retinal † Ganglion Cells in Mice
cells Article Intravitreal Co-Administration of GDNF and CNTF Confers Synergistic and Long-Lasting Protection against Injury-Induced Cell Death of Retinal y Ganglion Cells in Mice 1, 1, 1 2 2 Simon Dulz z , Mahmoud Bassal z, Kai Flachsbarth , Kristoffer Riecken , Boris Fehse , Stefanie Schlichting 1, Susanne Bartsch 1 and Udo Bartsch 1,* 1 Department of Ophthalmology, Experimental Ophthalmology, University Medical Center Hamburg-Eppendorf, 20246 Hamburg, Germany; [email protected] (S.D.); [email protected] (M.B.); kaifl[email protected] (K.F.); [email protected] (S.S.); [email protected] (S.B.) 2 Research Department Cell and Gene Therapy, University Medical Center Hamburg-Eppendorf, 20246 Hamburg, Germany; [email protected] (K.R.); [email protected] (B.F.) * Correspondence: [email protected]; Tel.: +49-40-7410-55945 A first draft of this manuscript is part of the unpublished doctoral thesis of Mahmoud Bassal. y Shared first authorship. z Received: 9 August 2020; Accepted: 9 September 2020; Published: 11 September 2020 Abstract: We have recently demonstrated that neural stem cell-based intravitreal co-administration of glial cell line-derived neurotrophic factor (GDNF) and ciliary neurotrophic factor (CNTF) confers profound protection to injured retinal ganglion cells (RGCs) in a mouse optic nerve crush model, resulting in the survival of ~38% RGCs two months after the nerve lesion. Here, we analyzed whether this neuroprotective effect is long-lasting and studied the impact of the pronounced RGC rescue on axonal regeneration. To this aim, we co-injected a GDNF- and a CNTF-overexpressing neural stem cell line into the vitreous cavity of adult mice one day after an optic nerve crush and determined the number of surviving RGCs 4, 6 and 8 months after the lesion. -

Proseek Multiplex Oncology I V296×96
Proseek Multiplex Oncology I v296×96 Adrenomedullin (AM) P35318 Fms-related tyrosine kinase 3 ligand (Flt3L) P49771 Amphiregulin (AR) P15514 Folate receptor alpha (FR-alpha) P15328 Angiopoietin-1 receptor (TIE2) Q02763 Follistatin (FS) P19883 B-cell activating factor (BAFF) Q9Y275 Furin (FUR) P09958 Cadherin-3 (CDH3) P22223 Growth hormone (GH) P01241 Carbonic anhydrase IX (CAIX) Q16790 Growth/differentiation factor 15 (GDF-15) Q99988 Carcinoembryonic antigen (CEA) P06731 Heparin-binding EGF-like growth factor (HB-EGF) Q99075 Caspase-3 (CASP-3) P42574 Hepatocyte growth factor (HGF) P14210 C-C motif chemokine 19 (CCL19) Q99731 ICOS ligand (ICOSLG) O75144 CD40 ligand (CD40-L) P29965 Immunoglobulin-like transcript 3 (ILT-3) Q8NHJ6 C-X-C motif chemokine 5 (CXCL5 ) P42830 Integrin alpha-1 (ITGA1) P56199 C-X-C motif chemokine 9 (CXCL9 ) Q07325 Interferon gamma (IFN-gamma) P01579 C-X-C motif chemokine 10 (CXCL10 ) P02778 Interleukin-1 receptor antagonist protein (IL-1ra) P18510 C-X-C motif chemokine 11 (CXCL11 ) O14625 Interleukin-2 (IL-2) P60568 C-X-C motif chemokine 13 (CXCL13 ) O43927 Interleukin-6 (IL-6) P05231 Cyclin-dependent kinase inhibitor 1 (CDKN1A) P38936 Interleukin-6 receptor subunit alpha (IL-6RA) P08887 Cystatin-B (CSTB) P04080 Interleukin-7 (IL-7) P13232 Early activation antigen CD69 (CD69 ) Q07108 Interleukin-8 (IL-8) P10145 Epidermal growth factor receptor (EGFR ) P00533 Interleukin-12 (IL-12) P29460; P29459 Epididymal secretory protein E4 (HE4 ) Q14508 Interleukin-17 receptor B (IL-17RB ) Q9NRM6 Epithelial cell adhesion molecule -

Effects of PACAP on Schwann Cells
International Journal of Molecular Sciences Review Effects of PACAP on Schwann Cells: Focus on Nerve Injury 1, 2, 1 3 Grazia Maugeri y, Agata Grazia D’Amico y, Giuseppe Musumeci , Dora Reglodi and Velia D’Agata 1,* 1 Department of Biomedical and Biotechnological Sciences, Section of Anatomy, Histology and Movement Sciences, University of Catania, 95100 Catania, Italy; [email protected] (G.M.); [email protected] (G.M.) 2 Department of Drug Sciences, University of Catania, 95100 Catania, Italy; [email protected] 3 Department of Anatomy, MTA-PTE PACAP Research Team, University of Pécs Medical School, Szigeti út 12, H-7624 Pécs, Hungary; [email protected] * Correspondence: [email protected]; Tel.: +39-095-3782039 These authors contributed equally to this work. y Received: 29 September 2020; Accepted: 2 November 2020; Published: 3 November 2020 Abstract: Schwann cells, the most abundant glial cells of the peripheral nervous system, represent the key players able to supply extracellular microenvironment for axonal regrowth and restoration of myelin sheaths on regenerating axons. Following nerve injury, Schwann cells respond adaptively to damage by acquiring a new phenotype. In particular, some of them localize in the distal stump to form the Bungner band, a regeneration track in the distal site of the injured nerve, whereas others produce cytokines involved in recruitment of macrophages infiltrating into the nerve damaged area for axonal and myelin debris clearance. Several neurotrophic factors, including pituitary adenylyl cyclase-activating peptide (PACAP), promote survival and axonal elongation of injured neurons. The present review summarizes the evidence existing in the literature demonstrating the autocrine and/or paracrine action exerted by PACAP to promote remyelination and ameliorate the peripheral nerve inflammatory response following nerve injury. -

Neurotrophins and Their Receptors: a Convergence Point for Many Signalling Pathways
REVIEWS NEUROTROPHINS AND THEIR RECEPTORS: A CONVERGENCE POINT FOR MANY SIGNALLING PATHWAYS Moses V.Chao The neurotrophins are a family of proteins that are essential for the development of the vertebrate nervous system. Each neurotrophin can signal through two different types of cell surface receptor — the Trk receptor tyrosine kinases and the p75 neurotrophin receptor. Given the wide range of activities that are now associated with neurotrophins, it is probable that additional regulatory events and signalling systems are involved. Here, I review recent findings that neurotrophins, in addition to promoting survival and differentiation, exert various effects through surprising interactions with other receptors and ion channels. 5,6 LONG-TERM POTENTIATION The era of growth factor research began fifty years ago receptor . Despite considerable progress in understand- (LTP).An enduring increase in with the discovery of nerve growth factor (NGF). Since ing the roles of these receptors, additional mechanisms the amplitude of excitatory then, the momentum to study the NGF — or neu- are needed to explain the many cellular and synaptic postsynaptic potentials as a rotrophin — family has never abated because of their interactions that occur between neurons. An emerging result of high-frequency (tetanic) stimulation of afferent continuous capacity to provide new insights into neural view is that neurotrophin receptors act as sensors for var- pathways. It is measured both as function; the influence of neurotrophins spans from ious extracellular and intracellular inputs, and several the amplitude of excitatory developmental neurobiology to neurodegenerative and new mechanisms have recently been put forward. Here, I postsynaptic potentials and as psychiatric disorders. In addition to their classic effects will consider several ways in which Trk and p75 receptors the magnitude of the postsynaptic-cell population on neuronal cell survival, neurotrophins can also regu- might account for the unique effects of neurotrophins spike. -

Canine TGF-Alpha ELISA Kit
Canine TGF-alpha ELISA Kit Catalog #: AYQ-E10339 (96 wells) User Manual This kit is designed to quantitatively detect the levels of Canine TGF-alpha in cell lysates, serum/ plasma and other suitable sample solution. Manufactured and Distributed by: AssaySolution 310 W Cummings Park, Woburn, MA, 01801, USA Phone: (617) 238-1396, Fax: (617) 380-0053 Email: [email protected] FOR RESEARCH USE ONLY. NOT FOR DIAGNOSTIC OR THERAPEUTIC PURPOSES Important notes Before using this product, please read this manual carefully; after reading the subsequent contents of this manual, please note the following specially: • The operation should be carried out in strict accordance with the provided instructions. • Store the unused strips in a sealed foil bag at 2-8°C. • Always avoid foaming when mixing or reconstituting protein solutions. • Pipette reagents and samples into the center of each well, avoid bubbles. • The samples should be transferred into the assay wells within 15 minutes of dilution. • We recommend that all standards, testing samples are tested in duplicate. • Using serial diluted sample is recommended for first test to get the best dilution factor. • If the blue color develops too light after 15 minutes incubation with the substrate, it may be appropriate to extend the incubation time (Do not over-develop). • Avoid cross-contamination by changing tips, using separate reservoirs for each reagent. • Avoid using the suction head without extensive wash. • Do not mix the reagents from different batches. • Stop Solution should be added in the same order of the Substrate Solution. • TMB developing agent is light-sensitive. Avoid prolonged exposure to the light. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................